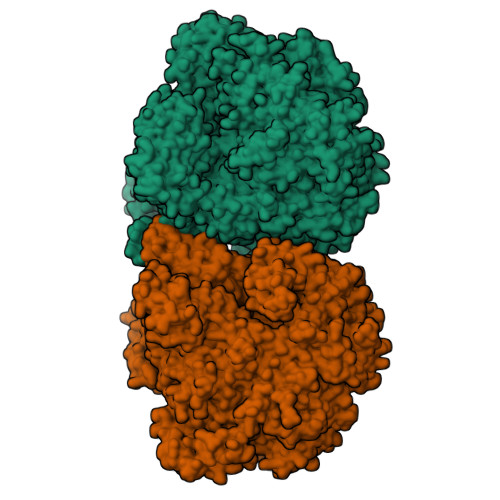
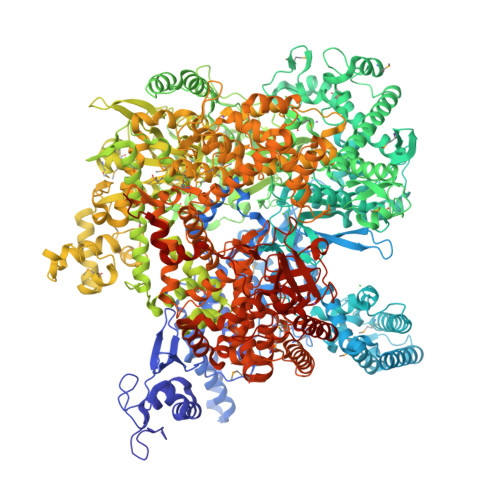
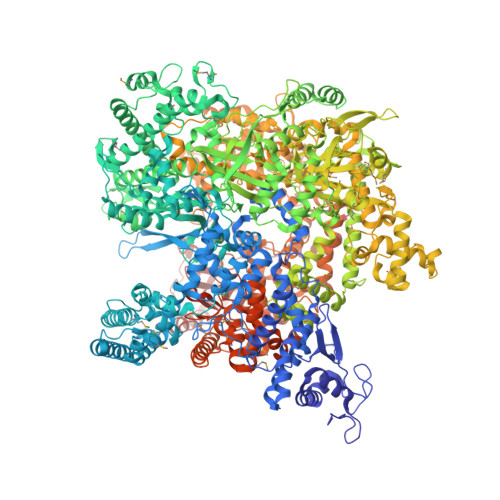
Structure and function of virion RNA polymerase of a crAss-like phage.
Drobysheva, A.V., Panafidina, S.A., Kolesnik, M.V., Klimuk, E.I., Minakhin, L., Yakunina, M.V., Borukhov, S., Nilsson, E., Holmfeldt, K., Yutin, N., Makarova, K.S., Koonin, E.V., Severinov, K.V., Leiman, P.G., Sokolova, M.L.(2020) Nature 589: 306-309
- PubMed: 33208949
- DOI: https://doi.org/10.1038/s41586-020-2921-5
- Primary Citation of Related Structures:
6VR4 - PubMed Abstract:
CrAss-like phages are a recently described expansive group of viruses that includes the most abundant virus in the human gut 1-3 . The genomes of all crAss-like phages encode a large virion-packaged protein 2,4 that contains a DFDxD sequence motif, which forms the catalytic site in cellular multisubunit RNA polymerases (RNAPs) 5 . Here, using Cellulophaga baltica crAss-like phage phi14:2 as a model system, we show that this protein is a DNA-dependent RNAP that is translocated into the host cell along with the phage DNA and transcribes early phage genes. We determined the crystal structure of this 2,180-residue enzyme in a self-inhibited state, which probably occurs before virion packaging. This conformation is attained with the help of a cleft-blocking domain that interacts with the active site and occupies the cavity in which the RNA-DNA hybrid binds. Structurally, phi14:2 RNAP is most similar to eukaryotic RNAPs that are involved in RNA interference 6,7 , although most of the phi14:2 RNAP structure (nearly 1,600 residues) maps to a new region of the protein fold space. Considering this structural similarity, we propose that eukaryal RNA interference polymerases have their origins in phage, which parallels the emergence of the mitochondrial transcription apparatus 8 .
Organizational Affiliation:
Center of Life Sciences, Skolkovo Institute of Science and Technology, Moscow, Russia.