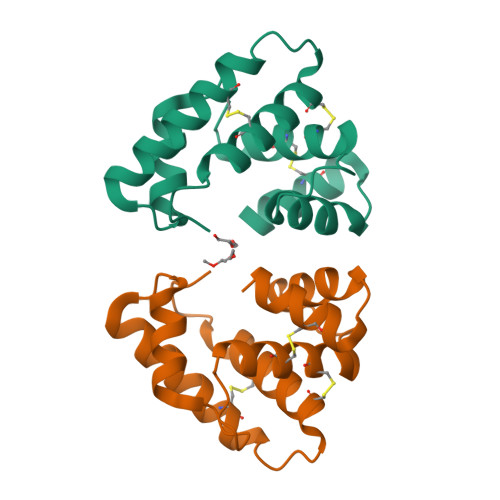
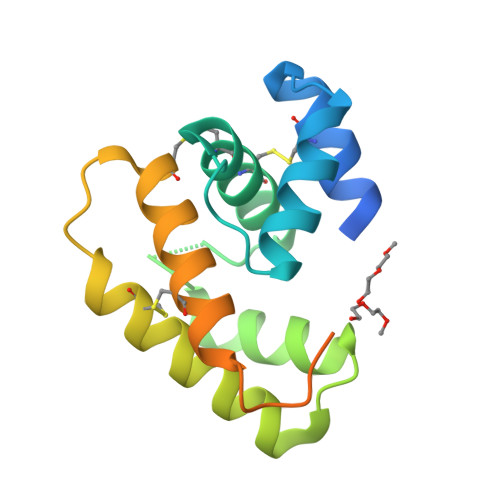
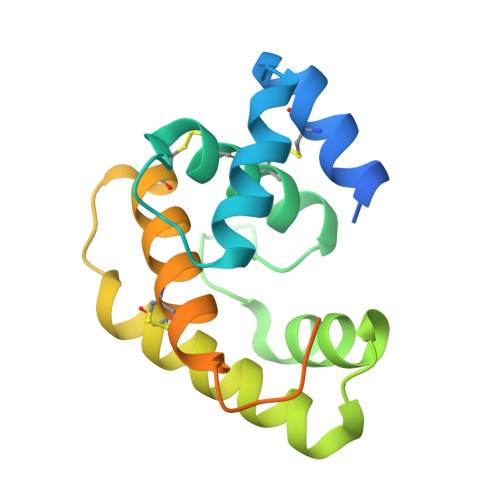
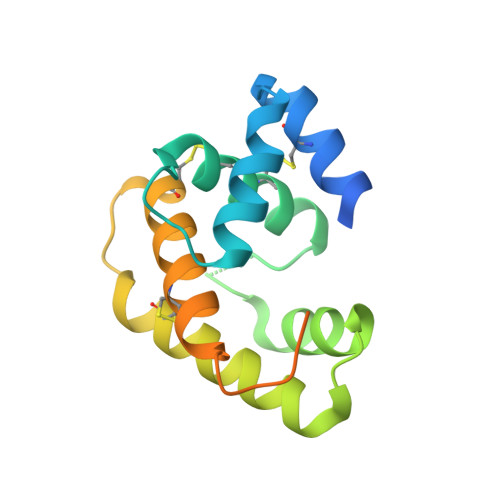
Crystal structure of Epiphyas postvittana pheromone binding protein 3.
Hamiaux, C., Carraher, C., Lofstedt, C., Corcoran, J.A.(2020) Sci Rep 10: 16366-16366
- PubMed: 33004932
- DOI: https://doi.org/10.1038/s41598-020-73294-8
- Primary Citation of Related Structures:
6VQ5 - PubMed Abstract:
The insect olfactory system operates as a well-choreographed ensemble of molecules which functions to selectively translate volatile chemical messages present in the environment into neuronal impulses that guide insect behaviour. Of these molecules, binding proteins are believed to transport hydrophobic odorant molecules across the aqueous lymph present in antennal sensilla to receptors present in olfactory sensory neurons. Though the exact mechanism through which these proteins operate is still under investigation, these carriers clearly play a critical role in determining what an insect can smell. Binding proteins that transport important sex pheromones are colloquially named pheromone binding proteins (PBPs). Here, we have produced a functional recombinant PBP from the horticultural pest, Epiphyas postvittana (EposPBP3), and experimentally solved its apo-structure through X-ray crystallography to a resolution of 2.60 Å. Structural comparisons with related lepidopteran PBPs further allowed us to propose models for the binding of pheromone components to EposPBP3. The data presented here represent the first structure of an olfactory-related protein from the tortricid family of moths, whose members cause billions of dollars in losses to agricultural producers each year. Knowledge of the structure of these important proteins will allow for subsequent studies in which novel, olfactory molecule-specific insecticides can be developed.
Organizational Affiliation:
The New Zealand Institute for Plant and Food Research Limited, Auckland, New Zealand.