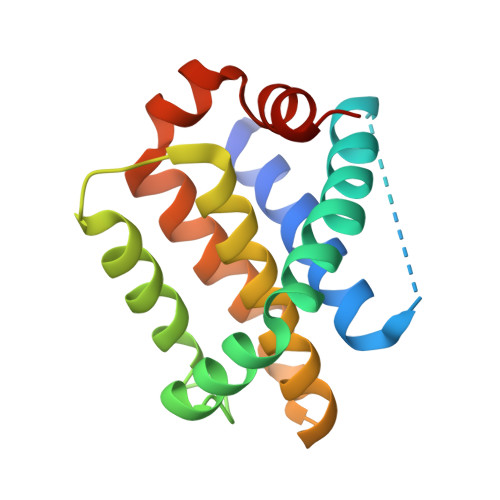
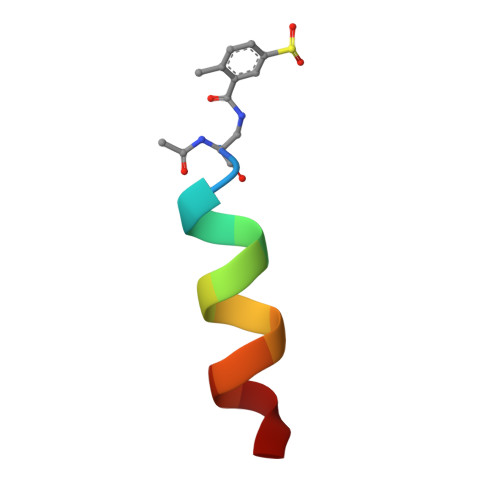
Design, Synthesis, and Structural Characterization of Lysine Covalent BH3 Peptides Targeting Mcl-1.
Gambini, L., Udompholkul, P., Baggio, C., Muralidharan, A., Kenjic, N., Assar, Z., Perry, J.J.P., Pellecchia, M.(2021) J Med Chem 64: 4903-4912
- PubMed: 33797903
- DOI: https://doi.org/10.1021/acs.jmedchem.1c00005
- Primary Citation of Related Structures:
6VBX - PubMed Abstract:
Modulating disease-relevant protein-protein interactions (PPIs) using pharmacological tools is a critical step toward the design of novel therapeutic strategies. Over the years, however, targeting PPIs has proven a very challenging task owing to the large interfacial areas. Our recent efforts identified possible novel routes for the design of potent and selective inhibitors of PPIs using a structure-based design of covalent inhibitors targeting Lys residues. In this present study, we report on the design, synthesis, and characterizations of the first Lys-covalent BH3 peptide that has a remarkable affinity and selectivity for hMcl-1 over the closely related hBfl-1 protein. Our structural studies, aided by X-ray crystallography, provide atomic-level details of the inhibitor interactions that can be used to further translate these discoveries into novel generation, Lys-covalent pro-apoptotic agents.
Organizational Affiliation:
Division of Biomedical Sciences, School of Medicine, University of California Riverside, 900 University Avenue, Riverside, California 92521, United States.