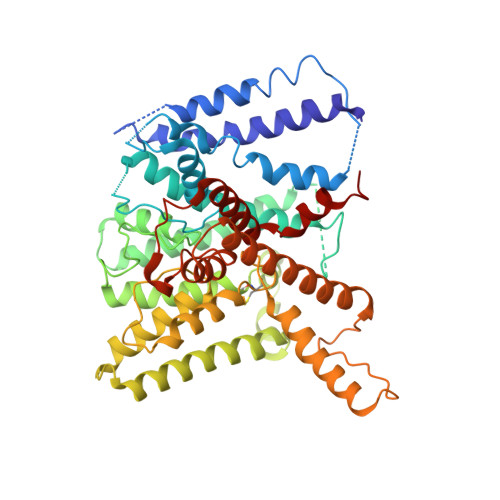
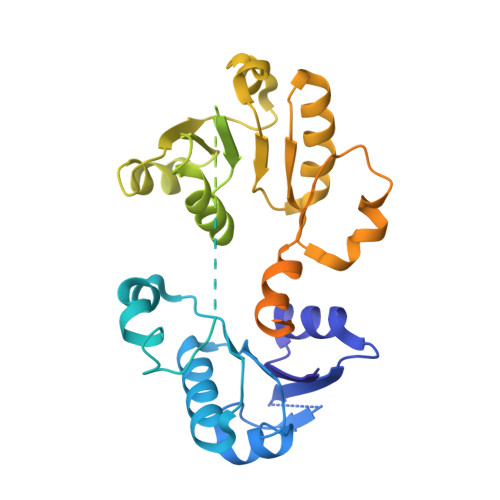
TrkA undergoes a tetramer-to-dimer conversion to open TrkH which enables changes in membrane potential.
Zhang, H., Pan, Y., Hu, L., Hudson, M.A., Hofstetter, K.S., Xu, Z., Rong, M., Wang, Z., Prasad, B.V.V., Lockless, S.W., Chiu, W., Zhou, M.(2020) Nat Commun 11: 547-547
- PubMed: 31992706
- DOI: https://doi.org/10.1038/s41467-019-14240-9
- Primary Citation of Related Structures:
6V4J, 6V4K, 6V4L - PubMed Abstract:
TrkH is a bacterial ion channel implicated in K + uptake and pH regulation. TrkH assembles with its regulatory protein, TrkA, which closes the channel when bound to ADP and opens it when bound to ATP. However, it is unknown how nucleotides control the gating of TrkH through TrkA. Here we report the structures of the TrkH-TrkA complex in the presence of ADP or ATP. TrkA forms a tetrameric ring when bound to ADP and constrains TrkH to a closed conformation. The TrkA ring splits into two TrkA dimers in the presence of ATP and releases the constraints on TrkH, resulting in an open channel conformation. Functional studies show that both the tetramer-to-dimer conversion of TrkA and the loss of constraints on TrkH are required for channel gating. In addition, deletion of TrkA in Escherichia coli depolarizes the cell, suggesting that the TrkH-TrkA complex couples changes in intracellular nucleotides to membrane potential.
Organizational Affiliation:
Verna and Marrs McLean Department of Biochemistry and Molecular Biology, Baylor College of Medicine, Houston, TX, 77030, USA.