Molecular characterization and three-dimensional structures of avian H8, H11, H14, H15 and swine H4 influenza virus hemagglutinins
Yang, H., Carney, P.J., Chang, J., Stevens, J.(2020) Heliyon 6: e04068
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
Currently 6V48 does not have a validation slider image.
(2020) Heliyon 6: e04068
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Hemagglutinin HA1 chain | 335 | Influenza A virus (A/mallard/Astrakhan/263/1982(H14N5)) | Mutation(s): 0 Gene Names: HA | 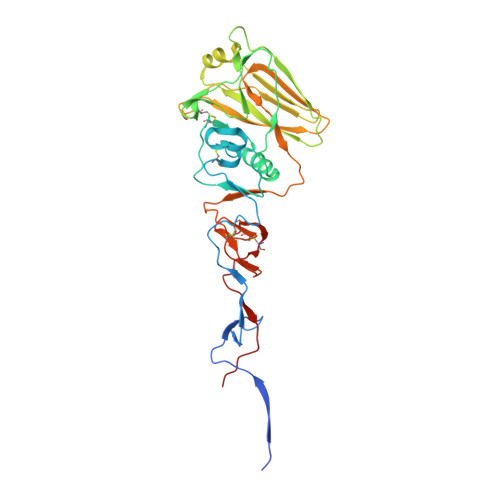 | |
UniProt | |||||
Find proteins for P26136 (Influenza A virus (strain A/Mallard/Astrakhan/263/1982 H14N5)) Explore P26136 Go to UniProtKB: P26136 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P26136 | ||||
Glycosylation | |||||
| Glycosylation Sites: 2 | |||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Hemagglutinin HA2 chain | 188 | Influenza A virus (A/mallard/Astrakhan/263/1982(H14N5)) | Mutation(s): 0 Gene Names: HA | 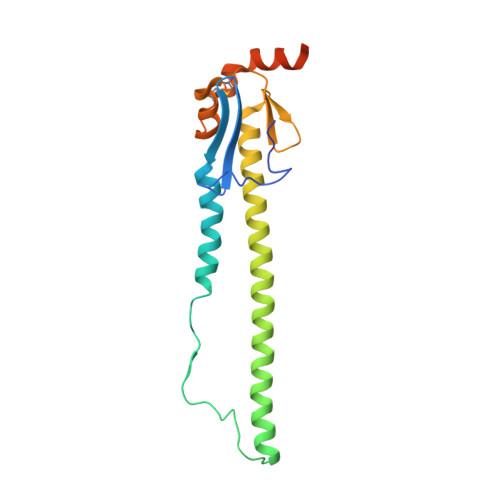 | |
UniProt | |||||
Find proteins for P26136 (Influenza A virus (strain A/Mallard/Astrakhan/263/1982 H14N5)) Explore P26136 Go to UniProtKB: P26136 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P26136 | ||||
Glycosylation | |||||
| Glycosylation Sites: 1 | |||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| NAG Query on NAG | AA [auth E] BA [auth G] CA [auth I] DA [auth K] Y [auth A] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 174.665 | α = 90 |
| b = 100.974 | β = 103.93 |
| c = 236.075 | γ = 90 |
| Software Name | Purpose |
|---|---|
| HKL-2000 | data scaling |
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| HKL-2000 | data reduction |
| PHASES | phasing |
Currently 6V48 does not have a validation slider image.