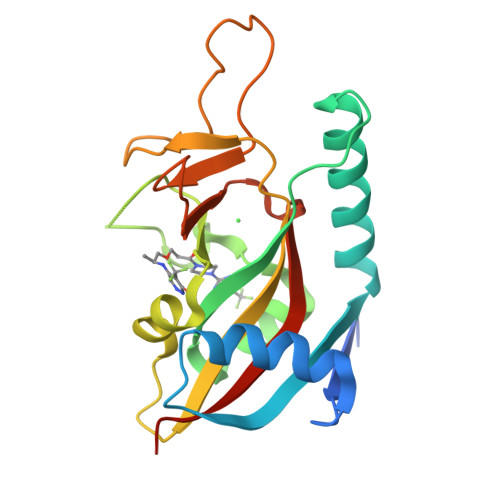
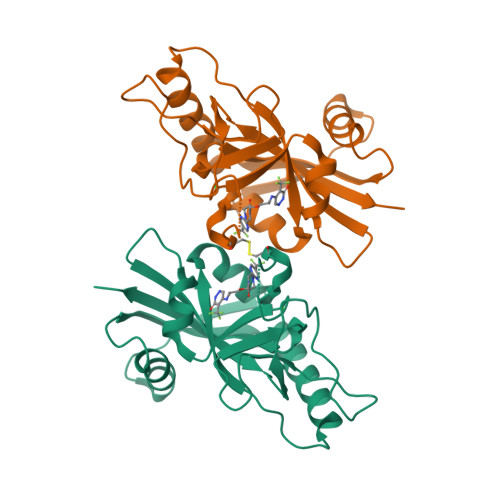
PARP7 negatively regulates the type I interferon response in cancer cells and its inhibition triggers antitumor immunity.
Gozgit, J.M., Vasbinder, M.M., Abo, R.P., Kunii, K., Kuplast-Barr, K.G., Gui, B., Lu, A.Z., Molina, J.R., Minissale, E., Swinger, K.K., Wigle, T.J., Blackwell, D.J., Majer, C.R., Ren, Y., Niepel, M., Varsamis, Z.A., Nayak, S.P., Bamberg, E., Mo, J.R., Church, W.D., Mady, A.S.A., Song, J., Utley, L., Rao, P.E., Mitchison, T.J., Kuntz, K.W., Richon, V.M., Keilhack, H.(2021) Cancer Cell 39: 1214-1226.e10
- PubMed: 34375612
- DOI: https://doi.org/10.1016/j.ccell.2021.06.018
- Primary Citation of Related Structures:
6V3W - PubMed Abstract:
PARP7 is a monoPARP that catalyzes the transfer of single units of ADP-ribose onto substrates to change their function. Here, we identify PARP7 as a negative regulator of nucleic acid sensing in tumor cells. Inhibition of PARP7 restores type I interferon (IFN) signaling responses to nucleic acids in tumor models. Restored signaling can directly inhibit cell proliferation and activate the immune system, both of which contribute to tumor regression. Oral dosing of the PARP7 small-molecule inhibitor, RBN-2397, results in complete tumor regression in a lung cancer xenograft and induces tumor-specific adaptive immune memory in an immunocompetent mouse cancer model, dependent on inducing type I IFN signaling in tumor cells. PARP7 is a therapeutic target whose inhibition induces both cancer cell-autonomous and immune stimulatory effects via enhanced IFN signaling. These data support the targeting of a monoPARP in cancer and introduce a potent and selective PARP7 inhibitor to enter clinical development.
Organizational Affiliation:
Ribon Therapeutics, 35 Cambridgepark Drive, Suite 300, Cambridge, MA 02140, USA. Electronic address: jgozgit@ribontx.com.