Cryo-EM structures of NPC1L1 reveal mechanisms of cholesterol transport and ezetimibe inhibition
Huang, C.S., Yu, X., Fordstrom, P., Choi, K., Chung, B.C., Roh, S.H., Chiu, W., Zhou, M., Min, X., Wang, Z.(2020) Sci Adv 6: eabb1989
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
(2020) Sci Adv 6: eabb1989
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| NPC1-like intracellular cholesterol transporter 1 | 1,336 | Rattus norvegicus | Mutation(s): 0 Gene Names: Npc1l1 Membrane Entity: Yes | 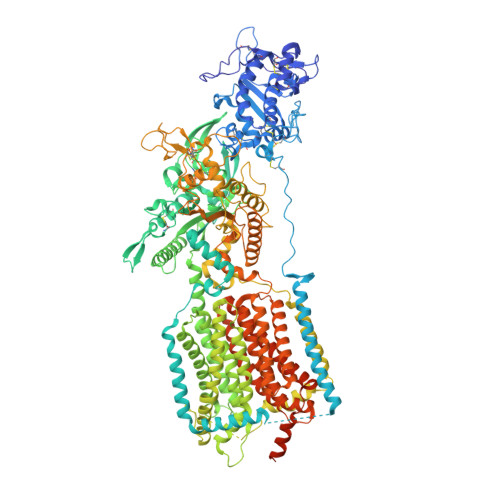 | |
UniProt | |||||
Find proteins for Q6T3U3 (Rattus norvegicus) Explore Q6T3U3 Go to UniProtKB: Q6T3U3 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q6T3U3 | ||||
Glycosylation | |||||
| Glycosylation Sites: 14 | Go to GlyGen: Q6T3U3-1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| CLR (Subject of Investigation/LOI) Query on CLR | P [auth A] | CHOLESTEROL C27 H46 O HVYWMOMLDIMFJA-DPAQBDIFSA-N |  | ||
| NAG Query on NAG | F [auth A] G [auth A] H [auth A] I [auth A] J [auth A] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| RECONSTRUCTION | RELION | 3 |