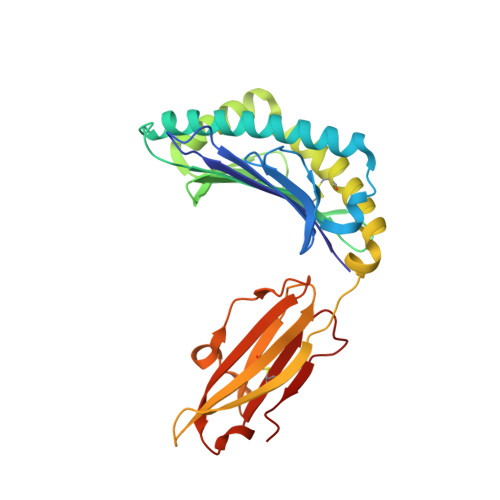
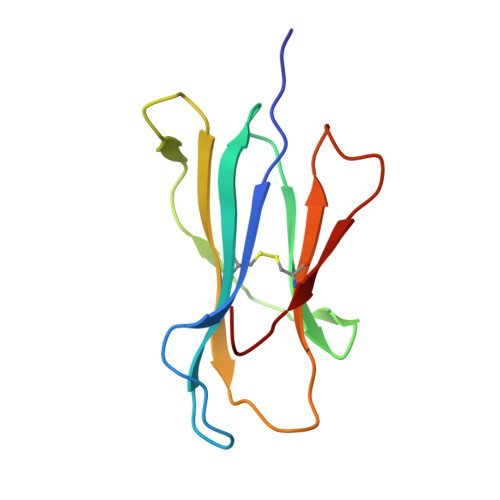
The molecular basis of how buried human leukocyte antigen polymorphism modulates natural killer cell function.
Saunders, P.M., MacLachlan, B.J., Pymm, P., Illing, P.T., Deng, Y., Wong, S.C., Oates, C.V.L., Purcell, A.W., Rossjohn, J., Vivian, J.P., Brooks, A.G.(2020) Proc Natl Acad Sci U S A 117: 11636-11647
- PubMed: 32404419
- DOI: https://doi.org/10.1073/pnas.1920570117
- Primary Citation of Related Structures:
6V2O, 6V2P, 6V2Q, 6V3J - PubMed Abstract:
Micropolymorphisms within human leukocyte antigen (HLA) class I molecules can change the architecture of the peptide-binding cleft, leading to differences in peptide presentation and T cell recognition. The impact of such HLA variation on natural killer (NK) cell recognition remains unclear. Given the differential association of HLA-B*57:01 and HLA-B*57:03 with the control of HIV, recognition of these HLA-B57 allomorphs by the killer cell immunoglobulin-like receptor (KIR) 3DL1 was compared. Despite differing by only two polymorphic residues, both buried within the peptide-binding cleft, HLA-B*57:01 more potently inhibited NK cell activation. Direct-binding studies showed KIR3DL1 to preferentially recognize HLA-B*57:01, particularly when presenting peptides with positively charged position (P)Ω-2 residues. In HLA-B*57:01, charged PΩ-2 residues were oriented toward the peptide-binding cleft and away from KIR3DL1. In HLA-B*57:03, the charged PΩ-2 residues protruded out from the cleft and directly impacted KIR3DL1 engagement. Accordingly, KIR3DL1 recognition of HLA class I ligands is modulated by both the peptide sequence and conformation, as determined by the HLA polymorphic framework, providing a rationale for understanding differences in clinical associations.
Organizational Affiliation:
Department of Microbiology and Immunology, Peter Doherty Institute for Infection and Immunity, The University of Melbourne, Melbourne, VIC 3010, Australia.