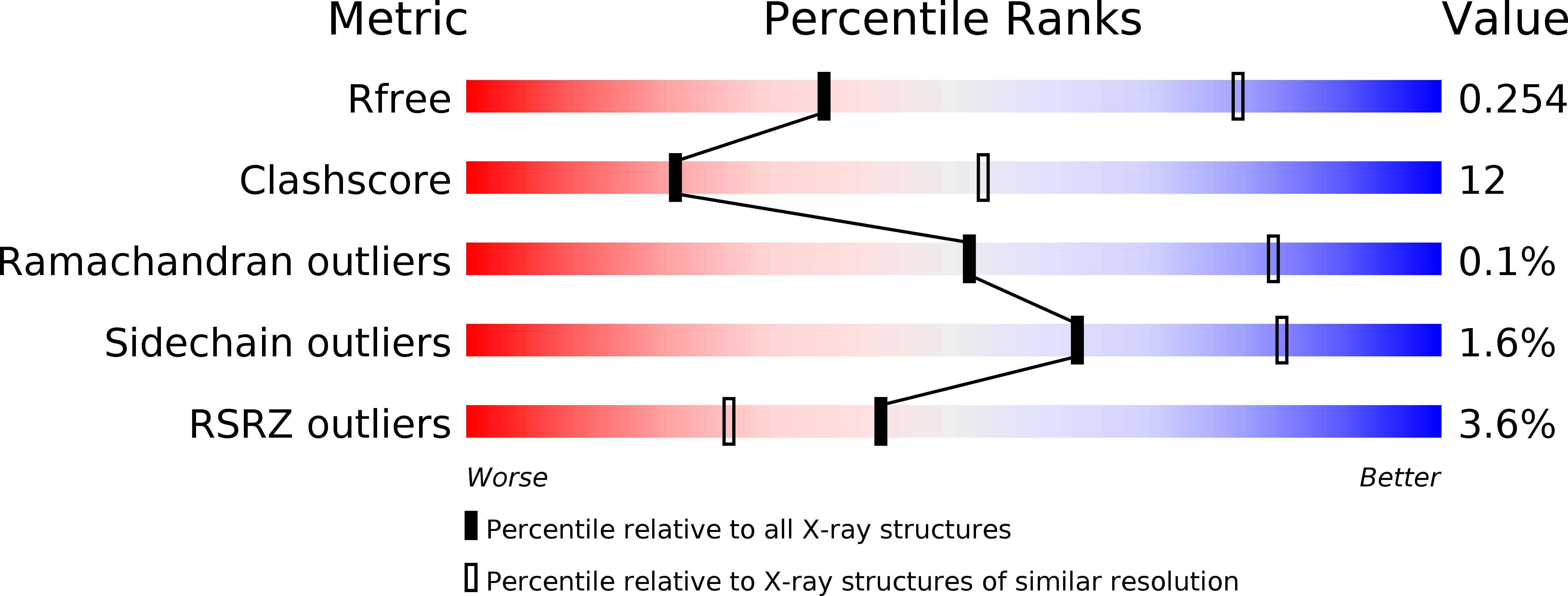
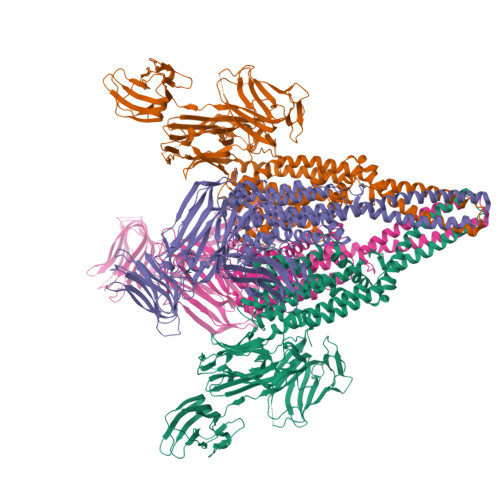
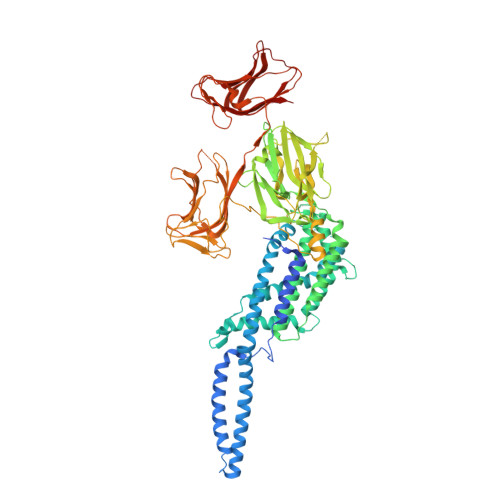
Crystal structure of a Vip3B family insecticidal protein reveals a new fold and a unique tetrameric assembly.
Zheng, M., Evdokimov, A.G., Moshiri, F., Lowder, C., Haas, J.(2020) Protein Sci 29: 824-829
- PubMed: 31840313
- DOI: https://doi.org/10.1002/pro.3803
- Primary Citation of Related Structures:
6V1V - PubMed Abstract:
Vegetatively expressed insecticidal proteins (VIPs) produced by Bacillus thuringiensis fall into several classes of which the third, VIP3, is known for their activity against several key Lepidopteran pests of commercial broad acre crops and because their mode of action does not overlap with that of crystalline insecticidal proteins. The details of the VIP3 structure and mode of action have remained obscure for the quarter century that has passed since their discovery. In the present article, we report the first crystal structure of a full-length VIP3 protein. Crystallization of this target required multiple rounds of construct optimization and screening-over 200 individual sequences were expressed and tested. This protein adopts a novel global fold that combines domains with hitherto unreported topology and containing elements seemingly borrowed from carbohydrate-binding domains, lectins, or from other insecticidal proteins.
Organizational Affiliation:
Protein Science Department, Bayer, Chesterfield, Missouri.