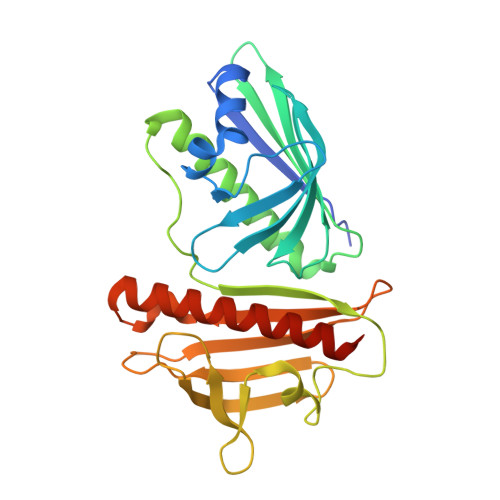
Structural characterization of DynU16, a START/Bet v1-like protein involved in dynemicin biosynthesis.
Alvarado, S.K., Miller, M.D., Bhardwaj, M., Thorson, J.S., Van Lanen, S.G., Phillips Jr., G.N.(2021) Acta Crystallogr F Struct Biol Commun 77: 328-333
- PubMed: 34605436
- DOI: https://doi.org/10.1107/S2053230X21008943
- Primary Citation of Related Structures:
6V04 - PubMed Abstract:
The 1.5 Å resolution crystal structure of DynU16, a protein identified in the dynemicin-biosynthetic gene cluster, is reported. The structure adopts a di-domain helix-grip fold with a uniquely positioned open cavity connecting the domains. The elongated dimensions of the cavity appear to be compatible with the geometry of a linear polyene, suggesting the involvement of DynU16 in the upstream steps of dynemicin biosynthesis.
Organizational Affiliation:
Department of BioSciences, Rice University, 6100 Main Street, Houston, TX 77005, USA.