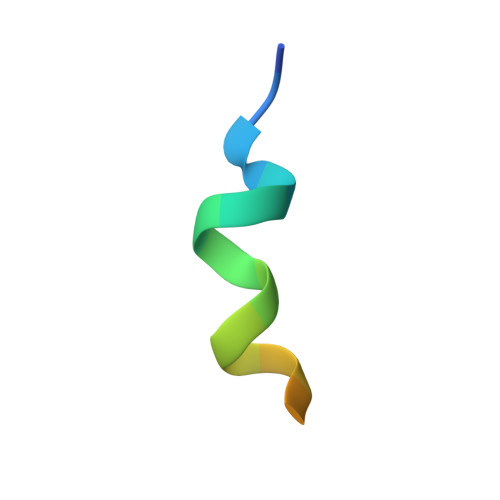
A high-affinity antibody against the CSP N-terminal domain lacks Plasmodium falciparum inhibitory activity.
Thai, E., Costa, G., Weyrich, A., Murugan, R., Oyen, D., Flores-Garcia, Y., Prieto, K., Bosch, A., Valleriani, A., Wu, N.C., Pholcharee, T., Scally, S.W., Wilson, I.A., Wardemann, H., Julien, J.P., Levashina, E.A.(2020) J Exp Med 217
- PubMed: 32790871
- DOI: https://doi.org/10.1084/jem.20200061
- Primary Citation of Related Structures:
6UUD - PubMed Abstract:
Malaria is a global health concern, and research efforts are ongoing to develop a superior vaccine to RTS,S/AS01. To guide immunogen design, we seek a comprehensive understanding of the protective humoral response against Plasmodium falciparum (Pf) circumsporozoite protein (PfCSP). In contrast to the well-studied responses to the repeat region and the C-terminus, the antibody response against the N-terminal domain of PfCSP (N-CSP) remains obscure. Here, we characterized the molecular recognition and functional efficacy of the N-CSP-specific monoclonal antibody 5D5. The crystal structure at 1.85-Å resolution revealed that 5D5 binds an α-helical epitope in N-CSP with high affinity through extensive shape and charge complementarity and the unusual utilization of an antibody N-linked glycan. Nevertheless, functional studies indicated low 5D5 binding to live Pf sporozoites and lack of sporozoite inhibition in vitro and in vivo. Overall, our data do not support the inclusion of the 5D5 N-CSP epitope into the next generation of CSP-based vaccines.
Organizational Affiliation:
Program in Molecular Medicine, The Hospital for Sick Children Research Institute, Toronto, Ontario, Canada.