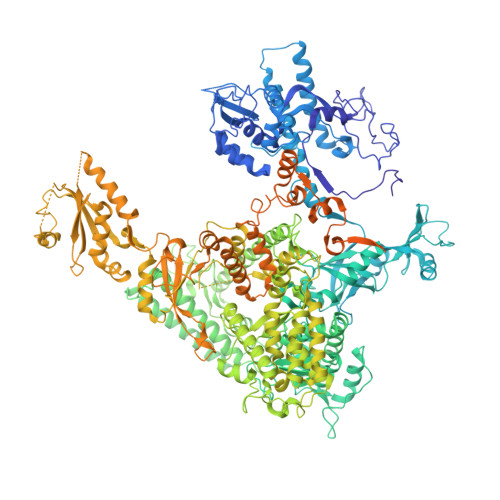
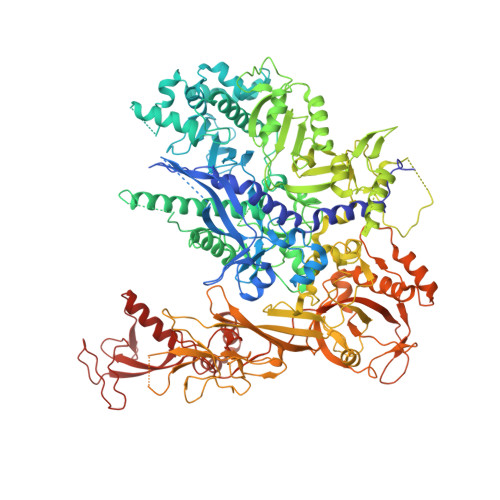
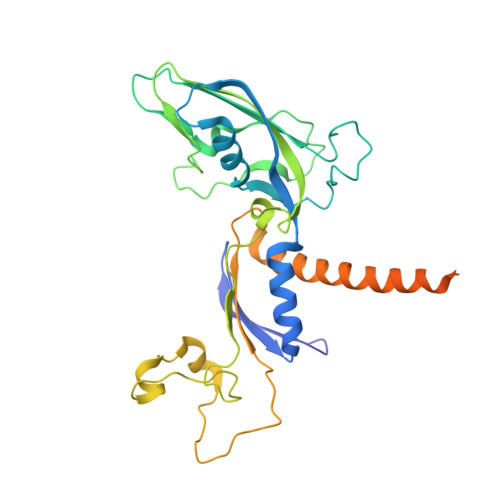
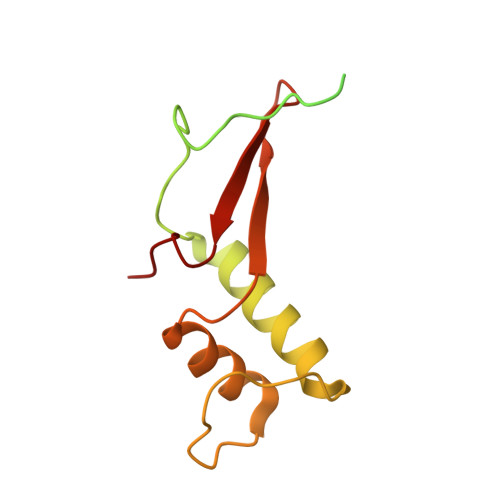
RNA polymerase II stalls on oxidative DNA damage via a torsion-latch mechanism involving lone pair-pi and CH-pi interactions.
Oh, J., Fleming, A.M., Xu, J., Chong, J., Burrows, C.J., Wang, D.(2020) Proc Natl Acad Sci U S A 117: 9338-9348
- PubMed: 32284409
- DOI: https://doi.org/10.1073/pnas.1919904117
- Primary Citation of Related Structures:
6UPX, 6UPY, 6UPZ, 6UQ0, 6UQ1, 6UQ2, 6UQ3 - PubMed Abstract:
Oxidation of guanine generates several types of DNA lesions, such as 8-oxoguanine (8OG), 5-guanidinohydantoin (Gh), and spiroiminodihydantoin (Sp). These guanine-derived oxidative DNA lesions interfere with both replication and transcription. However, the molecular mechanism of transcription processing of Gh and Sp remains unknown. In this study, by combining biochemical and structural analysis, we revealed distinct transcriptional processing of these chemically related oxidized lesions: 8OG allows both error-free and error-prone bypass, whereas Gh or Sp causes strong stalling and only allows slow error-prone incorporation of purines. Our structural studies provide snapshots of how polymerase II (Pol II) is stalled by a nonbulky Gh lesion in a stepwise manner, including the initial lesion encounter, ATP binding, ATP incorporation, jammed translocation, and arrested states. We show that while Gh can form hydrogen bonds with adenosine monophosphate (AMP) during incorporation, this base pair hydrogen bonding is not sufficient to hold an ATP substrate in the addition site and is not stable during Pol II translocation after the chemistry step. Intriguingly, we reveal a unique structural reconfiguration of the Gh lesion in which the hydantoin ring rotates ∼90° and is perpendicular to the upstream base pair planes. The perpendicular hydantoin ring of Gh is stabilized by noncanonical lone pair-π and CH-π interactions, as well as hydrogen bonds. As a result, the Gh lesion, as a functional mimic of a 1,2-intrastrand crosslink, occupies canonical -1 and +1 template positions and compromises the loading of the downstream template base. Furthermore, we suggest Gh and Sp lesions are potential targets of transcription-coupled repair.
Organizational Affiliation:
Division of Pharmaceutical Sciences, Skaggs School of Pharmacy and Pharmaceutical Sciences, University of California San Diego, La Jolla, CA 92093.