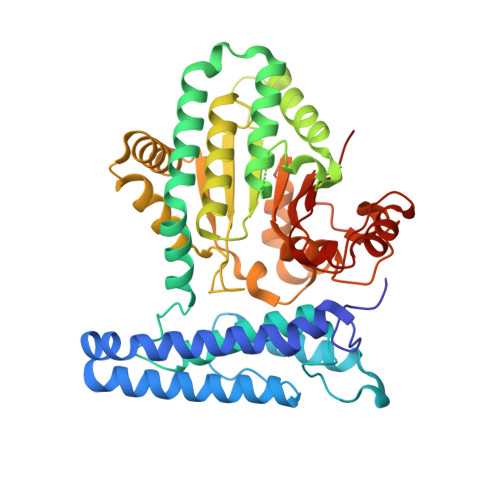
Human ARMT1 structure and substrate specificity indicates that it is a DUF89 family damage-control phosphatase.
Dennis, T.N., Kenjic, N., Kang, A.S., Lowenson, J.D., Kirkwood, J.S., Clarke, S.G., Perry, J.J.P.(2020) J Struct Biol 212: 107576-107576
- PubMed: 32682077
- DOI: https://doi.org/10.1016/j.jsb.2020.107576
- Primary Citation of Related Structures:
6UMQ, 6UMR - PubMed Abstract:
Metabolite damage control is a critical but poorly defined aspect of cellular biochemistry, which likely involves many of the so far functionally uncharacterized protein domain (domains of unknown function; DUFs). We have determined the crystal structure of the human DUF89 protein product of the C6ORF211 gene to 1.85 Å. The crystal structure shows that the protein contains a core α-β-α fold with an active site-bound metal ion and α-helical bundle N-terminal cap, which are both conserved features of subfamily III DUF89 domains. The biochemical activities of the human protein are conserved with those of a previously characterized budding yeast homolog, where an in vitro phosphatase activity is supported by divalent cations that include Co 2+ , Ni 2+ , Mn 2+ or Mg 2+ . Full steady-state kinetics parameters of human DUF89 using a standard PNPP phosphatase assay revealed a six times higher catalytic efficiency in presence of Co 2+ compared to Mg 2+ . The human enzyme targets a number of phosphate substrates similar to the budding yeast homolog, while it lacks a previously indicated methyltransferase activity. The highest activity on substrate was observed with fructose-1-phosphate, a potent glycating agent, and thus human DUF89 phosphatase activity may also play a role in limiting the buildup of phospho-glycan species and their related damaged metabolites.
Organizational Affiliation:
Department of Biochemistry, University of California, Riverside, Riverside, CA 92521, USA.