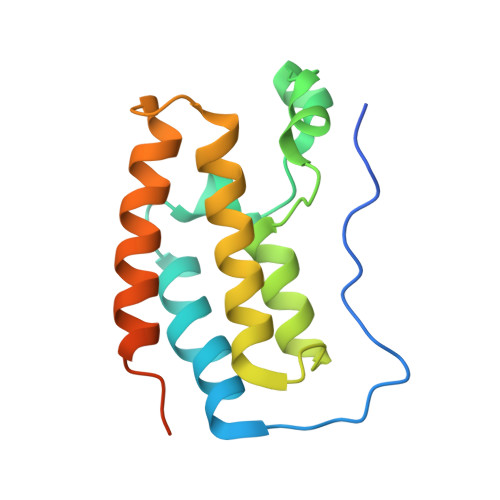
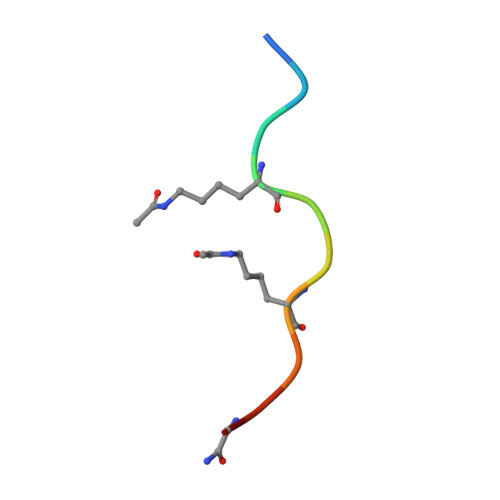
BET-Family Bromodomains Can Recognize Diacetylated Sequences from Transcription Factors Using a Conserved Mechanism.
Patel, K., Solomon, P.D., Walshe, J.L., Ford, D.J., Wilkinson-White, L., Payne, R.J., Low, J.K.K., Mackay, J.P.(2021) Biochemistry 60: 648-662
- PubMed: 33620209
- DOI: https://doi.org/10.1021/acs.biochem.0c00816
- Primary Citation of Related Structures:
6ULS - PubMed Abstract:
Almost all eukaryotic proteins receive diverse post-translational modifications (PTMs) that modulate protein activity. Many histone PTMs are well characterized, heavily influence gene regulation, and are often predictors of distinct transcriptional programs. Although our understanding of the histone PTM network has matured, much is yet to be understood about the roles of transcription factor (TF) PTMs, which might well represent a similarly complex and dynamic network of functional regulation. Members of the bromodomain and extra-terminal domain (BET) family of proteins recognize acetyllysine residues and relay the signals encoded by these modifications. Here, we have investigated the acetylation dependence of several functionally relevant BET-TF interactions in vitro using surface plasmon resonance, nuclear magnetic resonance, and X-ray crystallography. We show that motifs known to be acetylated in TFs E2F1 and MyoD1 can interact with all bromodomains of BRD2, BRD3, and BRD4. The interactions are dependent on diacetylation of the motifs and show a preference for the first BET bromodomain. Structural mapping of the interactions confirms a conserved mode of binding for the two TFs to the acetyllysine binding pocket of the BET bromodomains, mimicking that of other already established functionally important histone- and TF-BET interactions. We also examined a motif from the TF RelA that is known to be acetylated but were unable to observe any interaction, regardless of the acetylation state of the sequence. Our findings overall advance our understanding of BET-TF interactions and suggest a physical link between the important diacetylated motifs found in E2F1 and MyoD1 and the BET-family proteins.
Organizational Affiliation:
School of Life and Environmental Sciences, University of Sydney, Sydney, NSW 2006, Australia.