Structural basis of transmembrane coupling of the HIV-1 envelope glycoprotein.
Piai, A., Fu, Q., Cai, Y., Ghantous, F., Xiao, T., Shaik, M.M., Peng, H., Rits-Volloch, S., Chen, W., Seaman, M.S., Chen, B., Chou, J.J.(2020) Nat Commun 11: 2317-2317
- PubMed: 32385256
- DOI: https://doi.org/10.1038/s41467-020-16165-0
- Primary Citation of Related Structures:
6UJU, 6UJV - PubMed Abstract:
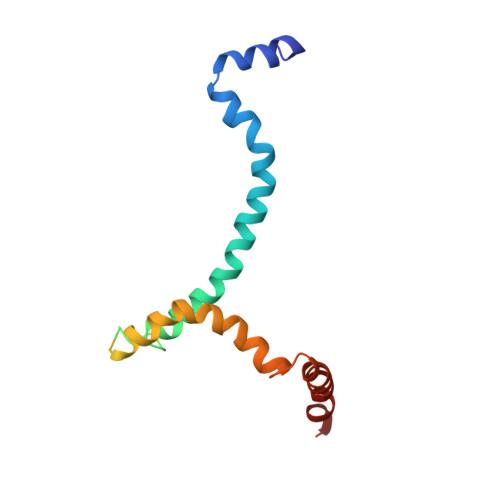
The prefusion conformation of HIV-1 envelope protein (Env) is recognized by most broadly neutralizing antibodies (bnAbs). Studies showed that alterations of its membrane-related components, including the transmembrane domain (TMD) and cytoplasmic tail (CT), can reshape the antigenic structure of the Env ectodomain. Using nuclear magnetic resonance (NMR) spectroscopy, we determine the structure of an Env segment encompassing the TMD and a large portion of the CT in bicelles. The structure reveals that the CT folds into amphipathic helices that wrap around the C-terminal end of the TMD, thereby forming a support baseplate for the rest of Env. NMR dynamics measurements provide evidences of dynamic coupling across the TMD between the ectodomain and CT. Pseudovirus-based neutralization assays suggest that CT-TMD interaction preferentially affects antigenic structure near the apex of the Env trimer. These results explain why the CT can modulate the Env antigenic properties and may facilitate HIV-1 Env-based vaccine design.
Organizational Affiliation:
Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, 250 Longwood Avenue, Boston, MA, 02115, USA.