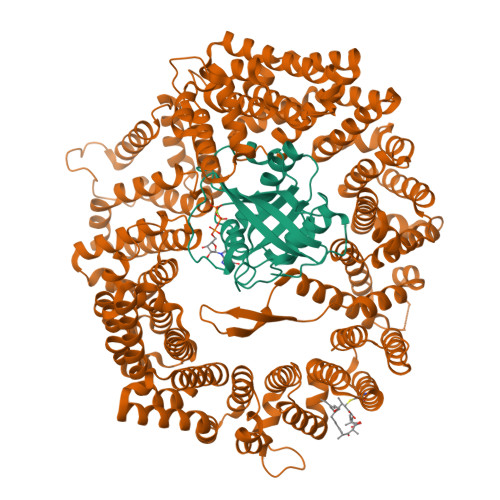
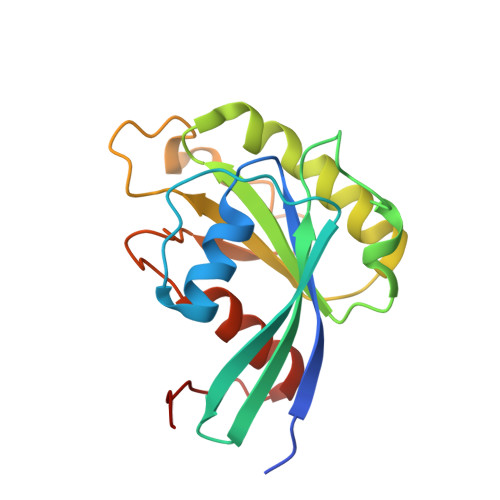
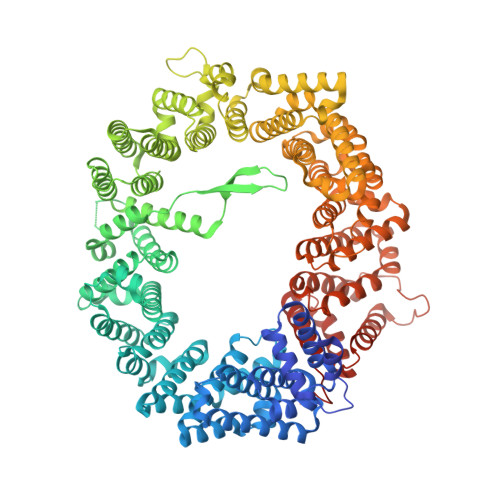
Characterization of Inhibition Reveals Distinctive Properties for Human andSaccharomyces cerevisiaeCRM1.
Shaikhqasem, A., Dickmanns, A., Neumann, P., Ficner, R.(2020) J Med Chem 63: 7545-7558
- PubMed: 32585100
- DOI: https://doi.org/10.1021/acs.jmedchem.0c00143
- Primary Citation of Related Structures:
6TVO - PubMed Abstract:
The receptor CRM1 is responsible for the nuclear export of many tumor-suppressor proteins and viral ribonucleoproteins. This renders CRM1 an interesting target for therapeutic intervention in diverse cancer types and viral diseases. Structural studies of Saccharomyces cerevisiae CRM1 ( Sc CRM1) complexes with inhibitors defined the molecular basis for CRM1 inhibition. Nevertheless, no structural information is available for inhibitors bound to human CRM1 ( Hs CRM1). Here, we present the structure of the natural inhibitor Leptomycin B bound to the Hs CRM1-RanGTP complex. Despite high sequence conservation and structural similarity in the NES-binding cleft region, Sc CRM1 exhibits 16-fold lower binding affinity than Hs CRM1 toward PKI-NES and significant differences in affinities toward potential CRM1 inhibitors. In contrast to Hs CRM1, competition assays revealed that a human adapted mutant Sc CRM1-T539C does not bind all inhibitors tested. Taken together, our data indicate the importance of using Hs CRM1 for molecular analysis and development of novel antitumor and antiviral drugs.
Organizational Affiliation:
Department of Molecular Structural Biology, Institute of Microbiology and Genetics, GZMB, Georg-August-University Göttingen, 37077 Göttingen, Germany.