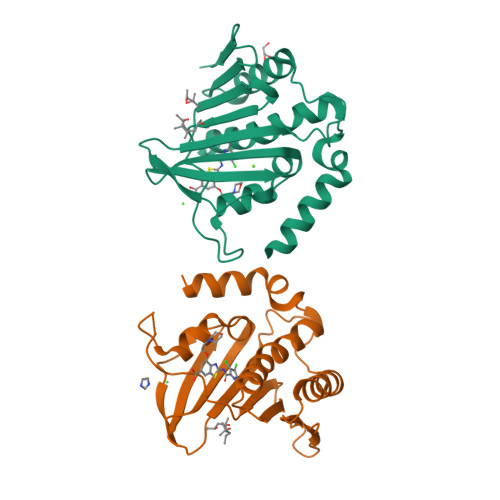
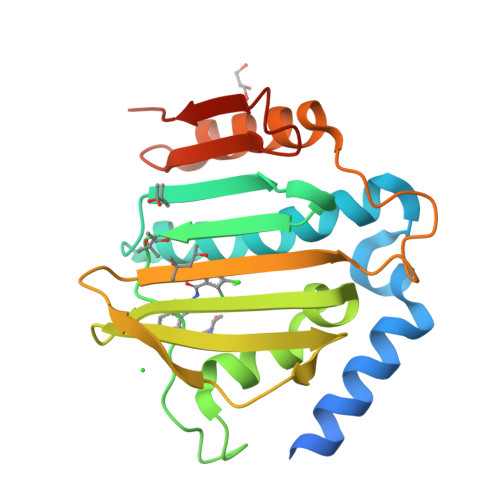
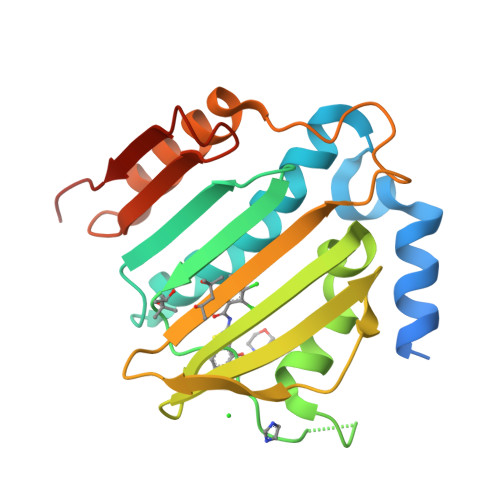
New dual ATP-competitive inhibitors of bacterial DNA gyrase and topoisomerase IV active against ESKAPE pathogens.
Durcik, M., Nyerges, A., Skok, Z., Skledar, D.G., Trontelj, J., Zidar, N., Ilas, J., Zega, A., Cruz, C.D., Tammela, P., Welin, M., Kimbung, Y.R., Focht, D., Benek, O., Revesz, T., Draskovits, G., Szili, P.E., Daruka, L., Pal, C., Kikelj, D., Masic, L.P., Tomasic, T.(2021) Eur J Med Chem 213: 113200-113200
- PubMed: 33524686
- DOI: https://doi.org/10.1016/j.ejmech.2021.113200
- Primary Citation of Related Structures:
6TTG - PubMed Abstract:
The rise in multidrug-resistant bacteria defines the need for identification of new antibacterial agents that are less prone to resistance acquisition. Compounds that simultaneously inhibit multiple bacterial targets are more likely to suppress the evolution of target-based resistance than monotargeting compounds. The structurally similar ATP binding sites of DNA gyrase and topoisomerase Ⅳ offer an opportunity to accomplish this goal. Here we present the design and structure-activity relationship analysis of balanced, low nanomolar inhibitors of bacterial DNA gyrase and topoisomerase IV that show potent antibacterial activities against the ESKAPE pathogens. For inhibitor 31c, a crystal structure in complex with Staphylococcus aureus DNA gyrase B was obtained that confirms the mode of action of these compounds. The best inhibitor, 31h, does not show any in vitro cytotoxicity and has excellent potency against Gram-positive (MICs: range, 0.0078-0.0625 μg/mL) and Gram-negative pathogens (MICs: range, 1-2 μg/mL). Furthermore, 31h inhibits GyrB mutants that can develop resistance to other drugs. Based on these data, we expect that structural derivatives of 31h will represent a step toward clinically efficacious multitargeting antimicrobials that are not impacted by existing antimicrobial resistance.
Organizational Affiliation:
University of Ljubljana, Faculty of Pharmacy, Aškerčeva cesta 7, 1000, Ljubljana, Slovenia.