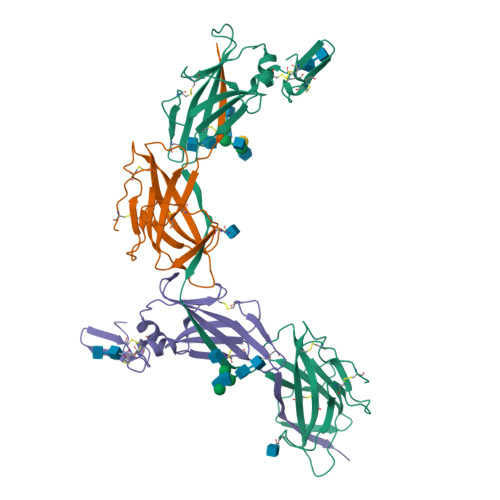
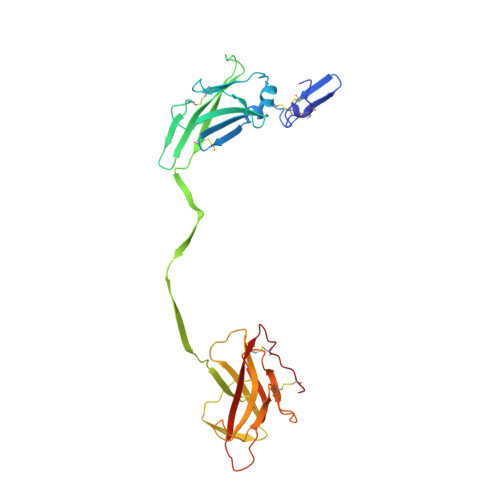
Cryo-EM structure of native human uromodulin, a zona pellucida module polymer.
Stsiapanava, A., Xu, C., Brunati, M., Zamora-Caballero, S., Schaeffer, C., Bokhove, M., Han, L., Hebert, H., Carroni, M., Yasumasu, S., Rampoldi, L., Wu, B., Jovine, L.(2020) EMBO J 39: e106807-e106807
- PubMed: 33196145
- DOI: https://doi.org/10.15252/embj.2020106807
- Primary Citation of Related Structures:
6TQK, 6TQL - PubMed Abstract:
Assembly of extracellular filaments and matrices mediating fundamental biological processes such as morphogenesis, hearing, fertilization, and antibacterial defense is driven by a ubiquitous polymerization module known as zona pellucida (ZP) "domain". Despite the conservation of this element from hydra to humans, no detailed information is available on the filamentous conformation of any ZP module protein. Here, we report a cryo-electron microscopy study of uromodulin (UMOD)/Tamm-Horsfall protein, the most abundant protein in human urine and an archetypal ZP module-containing molecule, in its mature homopolymeric state. UMOD forms a one-start helix with an unprecedented 180-degree twist between subunits enfolded by interdomain linkers that have completely reorganized as a result of propeptide dissociation. Lateral interaction between filaments in the urine generates sheets exposing a checkerboard of binding sites to capture uropathogenic bacteria, and UMOD-based models of heteromeric vertebrate egg coat filaments identify a common sperm-binding region at the interface between subunits.
Organizational Affiliation:
Department of Biosciences and Nutrition, Karolinska Institutet, Huddinge, Sweden.