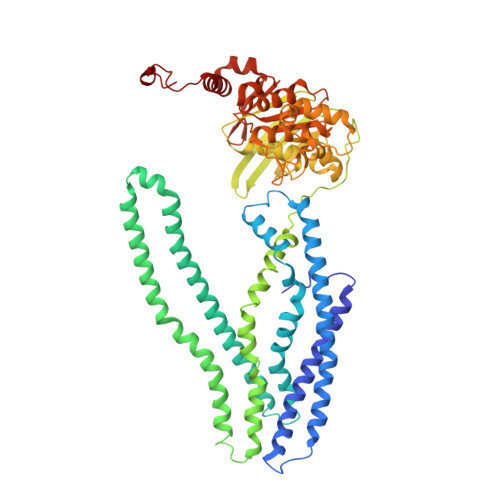
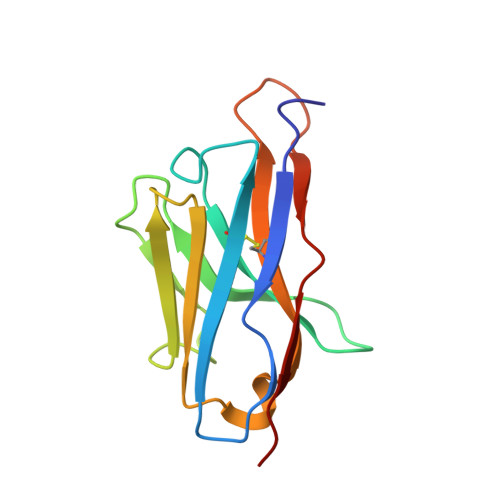
The ABC exporter IrtAB imports and reduces mycobacterial siderophores.
Arnold, F.M., Weber, M.S., Gonda, I., Gallenito, M.J., Adenau, S., Egloff, P., Zimmermann, I., Hutter, C.A.J., Hurlimann, L.M., Peters, E.E., Piel, J., Meloni, G., Medalia, O., Seeger, M.A.(2020) Nature 580: 413-417
- PubMed: 32296173
- DOI: https://doi.org/10.1038/s41586-020-2136-9
- Primary Citation of Related Structures:
6TEJ, 6TEK - PubMed Abstract:
Intracellular replication of the deadly pathogen Mycobacterium tuberculosis relies on the production of small organic molecules called siderophores that scavenge iron from host proteins 1 . M. tuberculosis produces two classes of siderophore, lipid-bound mycobactin and water-soluble carboxymycobactin 2,3 . Functional studies have revealed that iron-loaded carboxymycobactin is imported into the cytoplasm by the ATP binding cassette (ABC) transporter IrtAB 4 , which features an additional cytoplasmic siderophore interaction domain 5 . However, the predicted ABC exporter fold of IrtAB is seemingly contradictory to its import function. Here we show that membrane-reconstituted IrtAB is sufficient to import mycobactins, which are then reduced by the siderophore interaction domain to facilitate iron release. Structure determination by X-ray crystallography and cryo-electron microscopy not only confirms that IrtAB has an ABC exporter fold, but also reveals structural peculiarities at the transmembrane region of IrtAB that result in a partially collapsed inward-facing substrate-binding cavity. The siderophore interaction domain is positioned in close proximity to the inner membrane leaflet, enabling the reduction of membrane-inserted mycobactin. Enzymatic ATPase activity and in vivo growth assays show that IrtAB has a preference for mycobactin over carboxymycobactin as its substrate. Our study provides insights into an unusual ABC exporter that evolved as highly specialized siderophore-import machinery in mycobacteria.
Organizational Affiliation:
Institute of Medical Microbiology, University of Zurich, Zurich, Switzerland.