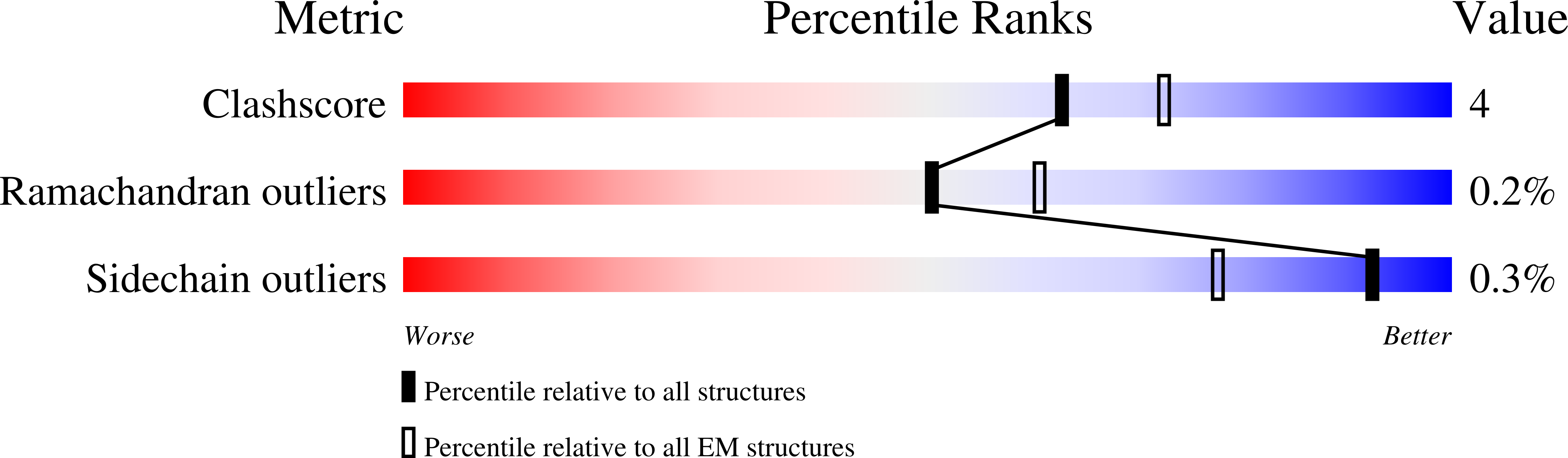
Antibiotic export by MexB multidrug efflux transporter is allosterically controlled by a MexA-OprM chaperone-like complex.
Glavier, M., Puvanendran, D., Salvador, D., Decossas, M., Phan, G., Garnier, C., Frezza, E., Cece, Q., Schoehn, G., Picard, M., Taveau, J.C., Daury, L., Broutin, I., Lambert, O.(2020) Nat Commun 11: 4948-4948
- PubMed: 33009415
- DOI: https://doi.org/10.1038/s41467-020-18770-5
- Primary Citation of Related Structures:
6T7S, 6TA5, 6TA6 - PubMed Abstract:
The tripartite multidrug efflux system MexAB-OprM is a major actor in Pseudomonas aeruginosa antibiotic resistance by exporting a large variety of antimicrobial compounds. Crystal structures of MexB and of its Escherichia coli homolog AcrB had revealed asymmetric trimers depicting a directional drug pathway by a conformational interconversion (from Loose and Tight binding pockets to Open gate (LTO) for drug exit). It remains unclear how MexB acquires its LTO form. Here by performing functional and cryo-EM structural investigations of MexB at various stages of the assembly process, we unveil that MexB inserted in lipid membrane is not set for active transport because it displays an inactive LTC form with a Closed exit gate. In the tripartite complex, OprM and MexA form a corset-like platform that converts MexB into the active form. Our findings shed new light on the resistance nodulation cell division (RND) cognate partners which act as allosteric factors eliciting the functional drug extrusion.
Organizational Affiliation:
University of Bordeaux, CBMN UMR 5248, Bordeaux INP, F-33600, Pessac, France.