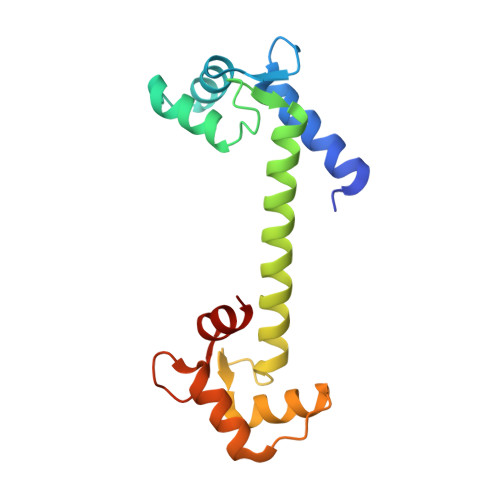
On the mechanism of calcium-dependent activation of NADPH oxidase 5 (NOX5).
Millana Fananas, E., Todesca, S., Sicorello, A., Masino, L., Pompach, P., Magnani, F., Pastore, A., Mattevi, A.(2020) FEBS J 287: 2486-2503
- PubMed: 31785178
- DOI: https://doi.org/10.1111/febs.15160
- Primary Citation of Related Structures:
6SZ5 - PubMed Abstract:
It is now accepted that reactive oxygen species (ROS) are not only dangerous oxidative agents but also chemical mediators of the redox cell signaling and innate immune response. A central role in ROS-controlled production is played by the NADPH oxidases (NOXs), a group of seven membrane-bound enzymes (NOX1-5 and DUOX1-2) whose unique function is to produce ROS. Here, we describe the regulation of NOX5, a widespread family member present in cyanobacteria, protists, plants, fungi, and the animal kingdom. We show that the calmodulin-like regulatory EF-domain of NOX5 is partially unfolded and detached from the rest of the protein in the absence of calcium. In the presence of calcium, the C-terminal lobe of the EF-domain acquires an ordered and more compact structure that enables its binding to the enzyme dehydrogenase (DH) domain. Our spectroscopic and mutagenesis studies further identified a set of conserved aspartate residues in the DH domain that are essential for NOX5 activation. Altogether, our work shows that calcium induces an unfolded-to-folded transition of the EF-domain that promotes direct interaction with a conserved regulatory region, resulting in NOX5 activation.
Organizational Affiliation:
Department of Biology and Biotechnology "Lazzaro Spallanzani", University of Pavia, Italy.