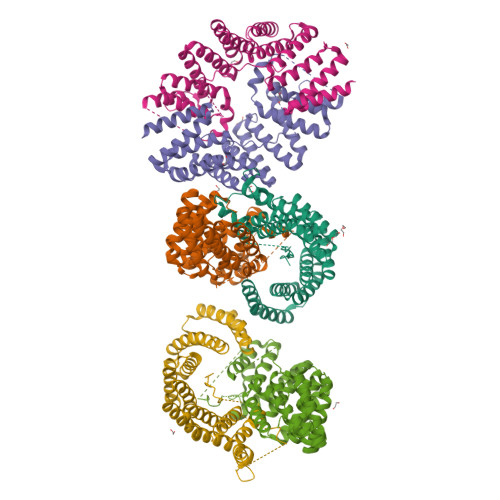
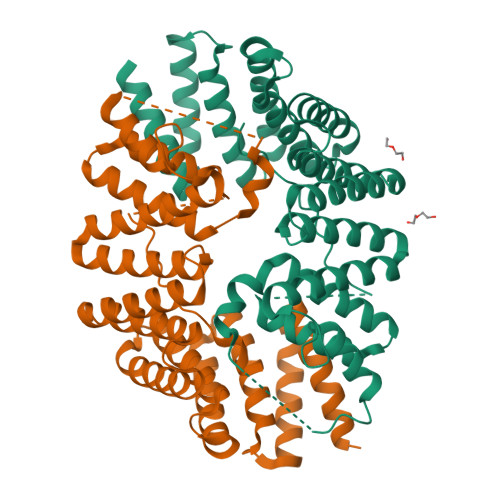
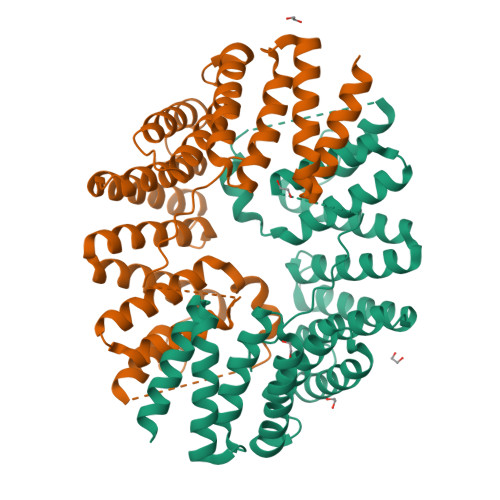
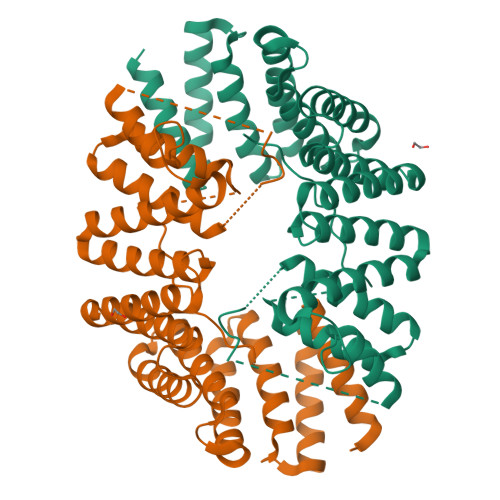
Fragment-linking peptide design yields a high-affinity ligand for microtubule-based transport.
Cross, J.A., Chegkazi, M.S., Steiner, R.A., Woolfson, D.N., Dodding, M.P.(2021) Cell Chem Biol 28: 1347
- PubMed: 33838110
- DOI: https://doi.org/10.1016/j.chembiol.2021.03.010
- Primary Citation of Related Structures:
6SWU - PubMed Abstract:
Synthetic peptides are attractive candidates to manipulate protein-protein interactions inside the cell as they mimic natural interactions to compete for binding. However, protein-peptide interactions are often dynamic and weak. A challenge is to design peptides that make improved interactions with the target. Here, we devise a fragment-linking strategy-"mash-up" design-to deliver a high-affinity ligand, KinTag, for the kinesin-1 motor. Using structural insights from natural micromolar-affinity cargo-adaptor ligands, we have identified and combined key binding features in a single, high-affinity ligand. An X-ray crystal structure demonstrates interactions as designed and reveals only a modest increase in interface area. Moreover, when genetically encoded, KinTag promotes transport of lysosomes with higher efficiency than natural sequences, revealing a direct link between motor-adaptor binding affinity and organelle transport. Together, these data demonstrate a fragment-linking strategy for peptide design and its application in a synthetic motor ligand to direct cellular cargo transport.
Organizational Affiliation:
School of Chemistry, University of Bristol, Cantock's Close, Bristol BS8 1TS, UK; School of Biochemistry, University of Bristol, University Walk, Bristol BS8 1TD, UK.