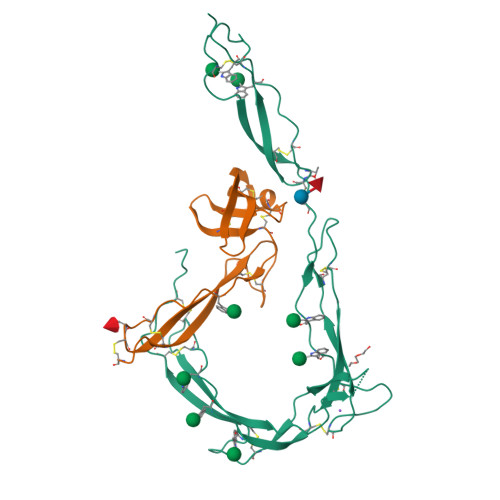
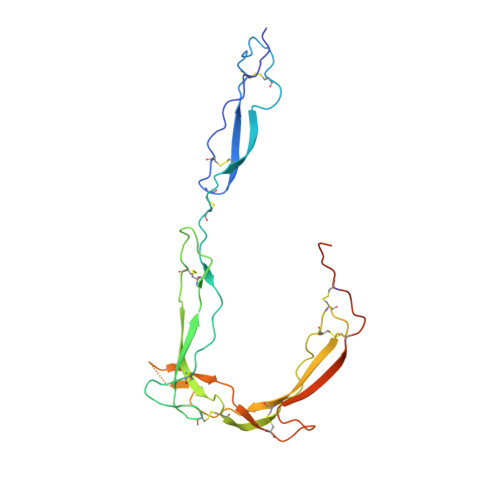
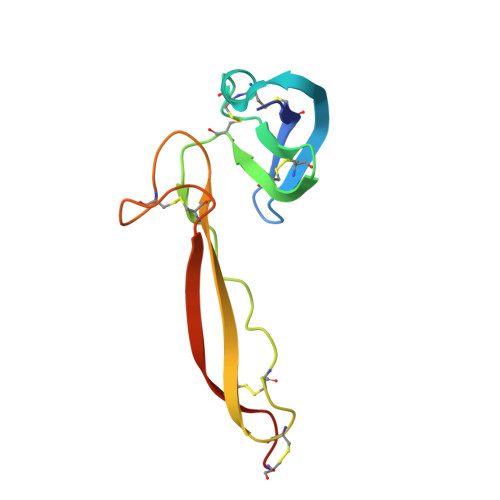
Insights Into Enhanced Complement Activation by Structures of Properdin and Its Complex With the C-Terminal Domain of C3b.
van den Bos, R.M., Pearce, N.M., Granneman, J., Brondijk, T.H.C., Gros, P.(2019) Front Immunol 10: 2097-2097
- PubMed: 31552043
- DOI: https://doi.org/10.3389/fimmu.2019.02097
- Primary Citation of Related Structures:
6S08, 6S0A, 6S0B - PubMed Abstract:
Properdin enhances complement-mediated opsonization of targeted cells and particles for immune clearance. Properdin occurs as dimers, trimers and tetramers in human plasma, which recognize C3b-deposited surfaces, promote formation, and prolong the lifetime of C3bBb-enzyme complexes that convert C3 into C3b, thereby enhancing the complement-amplification loop. Here, we report crystal structures of monomerized properdin, which was produced by co-expression of separate N- and C-terminal constructs that yielded monomer-sized properdin complexes that stabilized C3bBb. Consistent with previous low-resolution X-ray and EM data, the crystal structures revealed ring-shaped arrangements that are formed by interactions between thrombospondin type-I repeat (TSR) domains 4 and 6 of one protomer interacting with the N-terminal domain (which adopts a short transforming-growth factor B binding protein-like fold) and domain TSR1 of a second protomer, respectively. Next, a structure of monomerized properdin in complex with the C-terminal domain of C3b showed that properdin-domain TSR5 binds along the C-terminal α-helix of C3b, while two loops, one from domain TSR5 and one from TSR6, extend and fold around the C3b C-terminus like stirrups. This suggests a mechanistic model in which these TSR5 and TSR6 "stirrups" bridge interactions between C3b and factor B or its fragment Bb, and thereby enhance formation of C3bB pro-convertases and stabilize C3bBb convertases. In addition, properdin TSR6 would sterically block binding of the protease factor I to C3b, thus limiting C3b proteolytic degradation. The presence of a valine instead of a third tryptophan in the canonical Trp-ladder of TSR domains in TSR4 allows a remarkable ca. 60°-domain bending motion of TSR4. Together with variable positioning of TSR2 and, putatively, TSR3, this explains the conformational flexibility required for properdin to form dimers, trimers, and tetramers. In conclusion, the results indicate that binding avidity of oligomeric properdin is needed to distinguish surface-deposited C3b molecules from soluble C3b or C3 and suggest that properdin-mediated interactions bridging C3b-B and C3b-Bb enhance affinity, thus promoting convertase formation and stabilization. These mechanisms explain the enhancement of complement-mediated opsonization of targeted cells and particle for immune clearance.
Organizational Affiliation:
Crystal and Structural Chemistry, Department of Chemistry, Faculty of Science, Bijvoet Center for Biomolecular Research, Utrecht University, Utrecht, Netherlands.