Staphylococcal Complement Evasion Protein Sbi Stabilises C3d Dimers by Inducing an N-Terminal Helix Swap
Dunphy, R.W., Wahid, A.A., Back, C.R., Martin, R.L., Watts, A.G., Dodson, C.A., Crennell, S.J., van den Elsen, J.M.H.(2022) Front Immunol 13
Experimental Data Snapshot
Starting Model: experimental
View more details
wwPDB Validation 3D Report Full Report
(2022) Front Immunol 13
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Complement C3 | A, C [auth B] | 310 | Homo sapiens | Mutation(s): 0 Gene Names: C3, CPAMD1 | 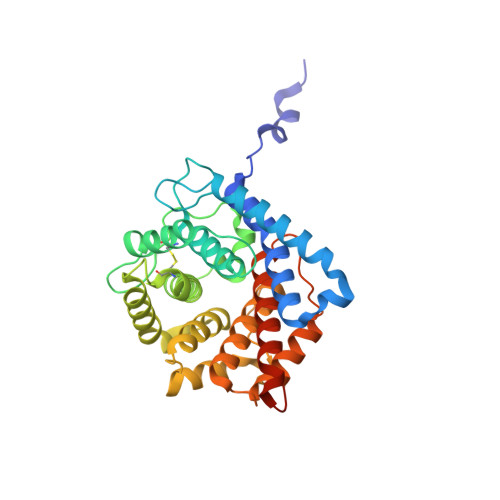 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P01024 (Homo sapiens) Explore P01024 Go to UniProtKB: P01024 | |||||
GTEx: ENSG00000125730 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P01024 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Immunoglobulin-binding protein Sbi | B [auth C], D | 81 | Staphylococcus aureus subsp. aureus Mu50 | Mutation(s): 0 Gene Names: sbi, SAV2418 |  |
UniProt | |||||
Find proteins for Q2FVK5 (Staphylococcus aureus (strain NCTC 8325 / PS 47)) Explore Q2FVK5 Go to UniProtKB: Q2FVK5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q2FVK5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| PEG Query on PEG | E [auth A] | DI(HYDROXYETHYL)ETHER C4 H10 O3 MTHSVFCYNBDYFN-UHFFFAOYSA-N |  | ||
| EDO Query on EDO | F [auth B] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 73.518 | α = 90 |
| b = 115.169 | β = 90 |
| c = 118.925 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHENIX | refinement |
| DIALS | data reduction |
| Aimless | data scaling |
| BALBES | phasing |
| Funding Organization | Location | Grant Number |
|---|---|---|
| Biotechnology and Biological Sciences Research Council | United Kingdom | BB/N022165/1 |