Novel beta- and gamma-Amino Acid-Derived Inhibitors of Prostate-Specific Membrane Antigen.
Kim, K., Kwon, H., Barinka, C., Motlova, L., Nam, S., Choi, D., Ha, H., Nam, H., Son, S.H., Minn, I., Pomper, M.G., Yang, X., Kutil, Z., Byun, Y.(2020) J Med Chem 63: 3261-3273
- PubMed: 32097010
- DOI: https://doi.org/10.1021/acs.jmedchem.9b02022
- Primary Citation of Related Structures:
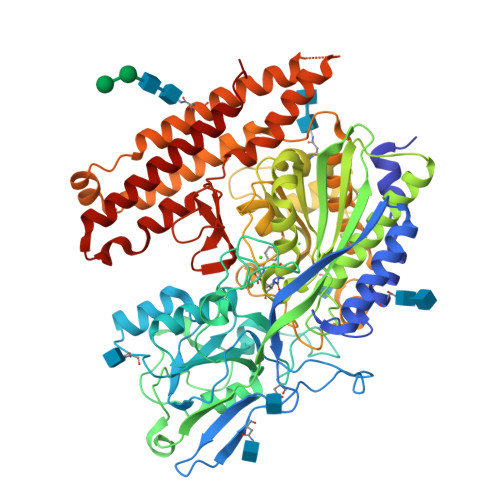
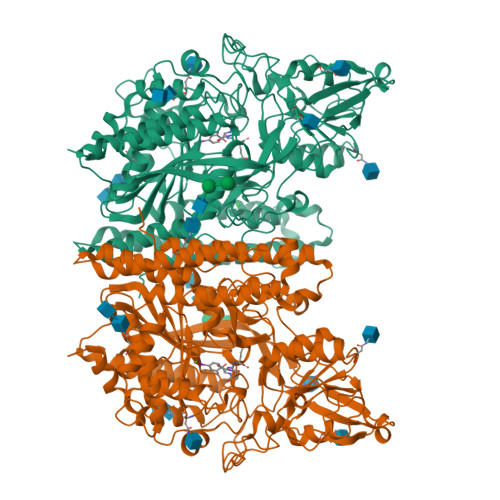
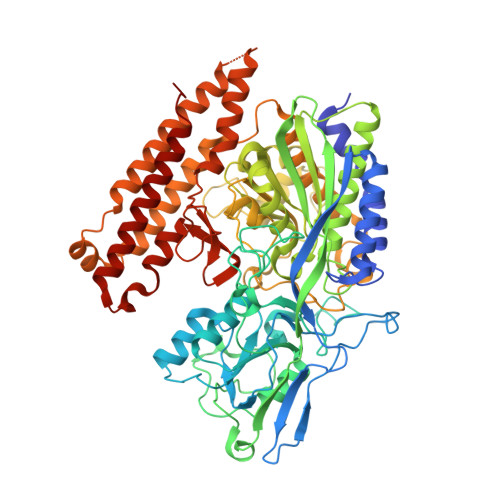
6RBC, 6S1X - PubMed Abstract:
Prostate-specific membrane antigen (PSMA) is an excellent biomarker for the early diagnosis of prostate cancer progression and metastasis. The most promising PSMA-targeted agents in the clinical phase are based on the Lys-urea-Glu motif, in which Lys and Glu are α-(l)-amino acids. In this study, we aimed to determine the effect of β- and γ-amino acids in the S1 pocket on the binding affinity for PSMA. We synthesized and evaluated the β- and γ-amino acid analogues with ( S )- or ( R )-configuration with keeping α-(l)-Glu as the S1'-binding pharmacophore. The structure-activity relationship studies identified that compound 13c , a β-amino acid analogue with ( R )-configuration, exhibited the most potent PSMA inhibitory activity with an IC 50 value of 3.97 nM. The X-ray crystal structure of PSMA in complex with 13c provided a mechanistic basis for the stereochemical preference of PSMA, which can guide the development of future PSMA inhibitors.
Organizational Affiliation:
College of Pharmacy, Korea University, 2511 Sejong-ro, Jochiwon-eup, Sejong 30019, Republic of Korea.