Binding and transport of D-aspartate by the glutamate transporter homolog Glt Tk .
Arkhipova, V., Trinco, G., Ettema, T.W., Jensen, S., Slotboom, D.J., Guskov, A.(2019) Elife 8
- PubMed: 30969168
- DOI: https://doi.org/10.7554/eLife.45286
- Primary Citation of Related Structures:
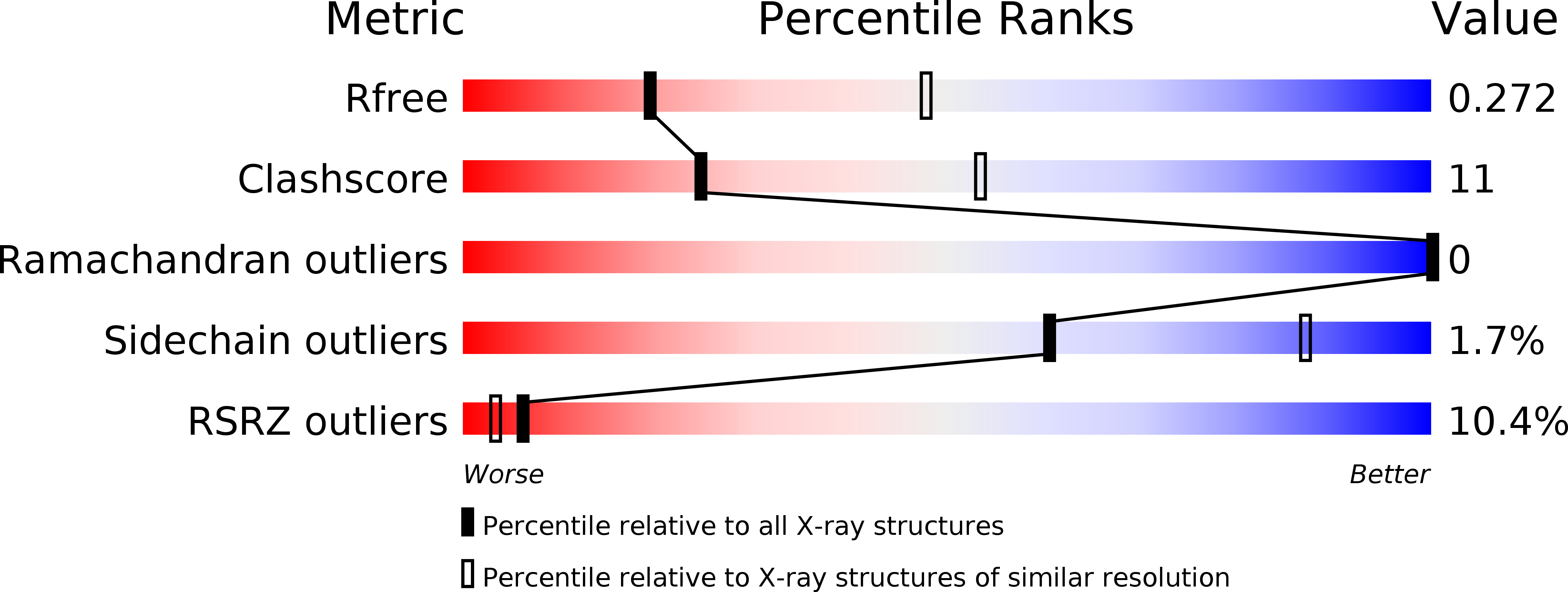
6R7R - PubMed Abstract:
Mammalian glutamate transporters are crucial players in neuronal communication as they perform neurotransmitter reuptake from the synaptic cleft. Besides L-glutamate and L-aspartate, they also recognize D-aspartate, which might participate in mammalian neurotransmission and/or neuromodulation. Much of the mechanistic insight in glutamate transport comes from studies of the archeal homologs Glt Ph from Pyrococcus horikoshii and Glt Tk from Thermococcus kodakarensis . Here, we show that Glt Tk transports D-aspartate with identical Na + : substrate coupling stoichiometry as L-aspartate, and that the affinities ( K d and K m ) for the two substrates are similar. We determined a crystal structure of Glt Tk with bound D-aspartate at 2.8 Å resolution. Comparison of the L- and D-aspartate bound Glt Tk structures revealed that D-aspartate is accommodated with only minor rearrangements in the structure of the binding site. The structure explains how the geometrically different molecules L- and D-aspartate are recognized and transported by the protein in the same way.
Organizational Affiliation:
Groningen Biomolecular Sciences and Biotechnology Institute, Zernike Institute for Advanced Materials, University of Groningen, Groningen, The Netherlands.