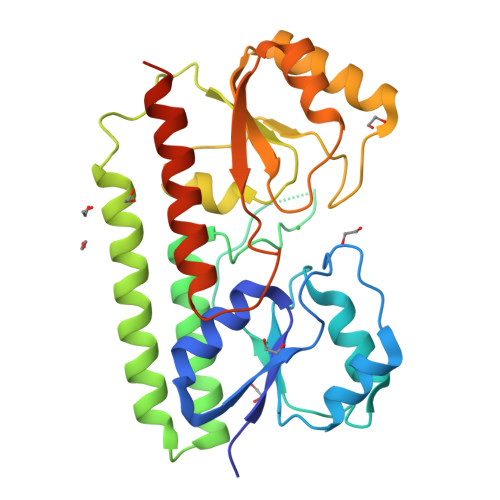
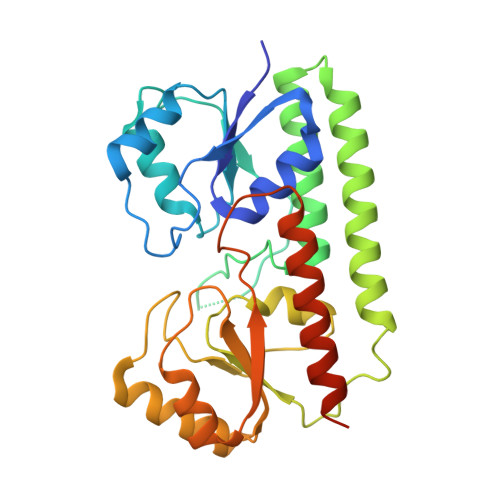
A unique ferrous iron binding mode is associated with large conformational changes for the transport protein FpvC of Pseudomonas aeruginosa.
Vigouroux, A., Aumont-Nicaise, M., Boussac, A., Marty, L., Lo Bello, L., Legrand, P., Brillet, K., Schalk, I.J., Morera, S.(2020) FEBS J 287: 295-309
- PubMed: 31318478
- DOI: https://doi.org/10.1111/febs.15004
- Primary Citation of Related Structures:
6R3Z, 6R44, 6R5S, 6R6K, 6RU4 - PubMed Abstract:
Pseudomonas aeruginosa secretes pyoverdine, a major siderophore to get access to iron, an essential nutrient. Pyoverdine scavenges ferric iron in the bacterial environment with the resulting complex internalized by bacteria. Releasing of iron from pyoverdine in the periplasm involves an iron reduction by an inner membrane reductase and two solute-binding proteins (SBPs) FpvC and FpvF in association with their ABC transporter. FpvC and FpvF belong to two different subgroups of SBPs within the structural cluster A: FpvC and FpvF were proposed to be a metal-binding protein and a ferrisiderophore-binding protein respectively. Here, we report the redox state and the binding mode of iron to FpvC. We first solved the crystal structure of FpvC bound to a fortuitous Ni 2+ by single anomalous dispersion method. Using a different protein purification strategy, we determined the structure of FpvC with manganese and iron, which binds to FpvC in a ferrous state as demonstrated by electron paramagnetic resonance. FpvC is the first example of a hexahistidine metal site among SBPs in which the Fe 2+ redox state is stabilized under aerobic conditions. Using biophysics methods, we showed that FpvC reversibly bind to a broad range of divalent ions. The structure of a mutant mimicking the apo FpvC reveals a protein in an open state with large conformational changes when compared with the metal-bound FpvC. These results highlight that the canonical metal site in FpvC is distinct from those yet described in SBPs and they provide new insights into the mechanism of PVD-Fe dissociation in P. aeruginosa.
Organizational Affiliation:
Institute for Integrative Biology of the Cell (I2BC), CNRS CEA Univ. Paris-Sud, Université Paris-Saclay, Gif-sur-Yvette, France.