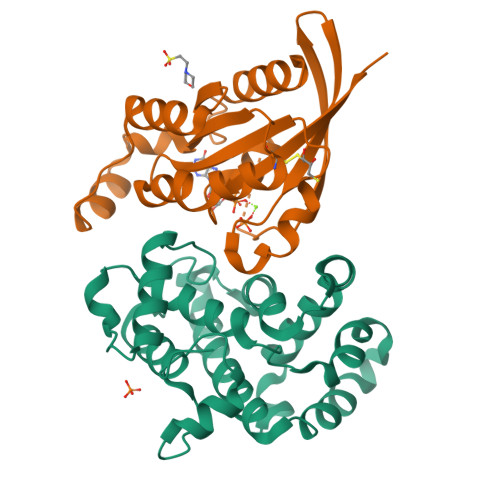
A GAP-GTPase-GDP-PiIntermediate Crystal Structure Analyzed by DFT Shows GTP Hydrolysis Involves Serial Proton Transfers.
Molt Jr., R.W., Pellegrini, E., Jin, Y.(2019) Chemistry 25: 8484-8488
- PubMed: 31038818
- DOI: https://doi.org/10.1002/chem.201901627
- Primary Citation of Related Structures:
6R3V - PubMed Abstract:
Cell signaling by small G proteins uses an ON to OFF signal based on conformational changes following the hydrolysis of GTP to GDP and release of dihydrogen phosphate (P i ). The catalytic mechanism of GTP hydrolysis by RhoA is strongly accelerated by a GAP protein and is now well defined, but timing of inorganic phosphate release and signal change remains unresolved. We have generated a quaternary complex for RhoA-GAP-GDP-P i . Its 1.75 Å crystal structure shows geometry for ionic and hydrogen bond coordination of GDP and P i in an intermediate state. It enables the selection of a QM core for DFT exploration of a 20 H-bonded network. This identifies serial locations of the two mobile protons from the original nucleophilic water molecule, showing how they move in three rational steps to form a stable quaternary complex. It also suggests how two additional proton transfer steps can facilitate P i release.
Organizational Affiliation:
Department of Biochemistry & Molecular Biology, Indiana University School of Medicine, Indianapolis, Indiana, 46202, USA.