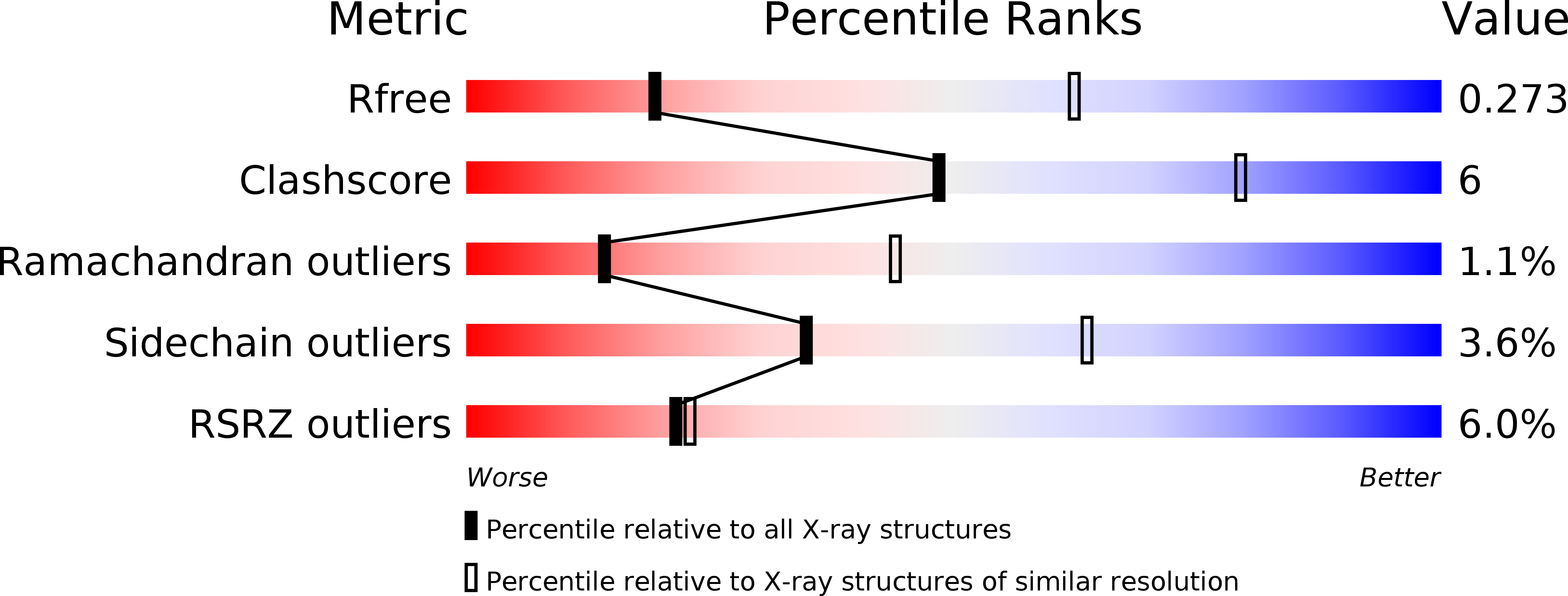
Structures of T7 bacteriophage portal and tail suggest a viral DNA retention and ejection mechanism.
Cuervo, A., Fabrega-Ferrer, M., Machon, C., Conesa, J.J., Fernandez, F.J., Perez-Luque, R., Perez-Ruiz, M., Pous, J., Vega, M.C., Carrascosa, J.L., Coll, M.(2019) Nat Commun 10: 3746-3746
- PubMed: 31431626
- DOI: https://doi.org/10.1038/s41467-019-11705-9
- Primary Citation of Related Structures:
6QWP, 6QX5, 6QXM, 6R21 - PubMed Abstract:
Double-stranded DNA bacteriophages package their genome at high pressure inside a procapsid through the portal, an oligomeric ring protein located at a unique capsid vertex. Once the DNA has been packaged, the tail components assemble on the portal to render the mature infective virion. The tail tightly seals the ejection conduit until infection, when its interaction with the host membrane triggers the opening of the channel and the viral genome is delivered to the host cell. Using high-resolution cryo-electron microscopy and X-ray crystallography, here we describe various structures of the T7 bacteriophage portal and fiber-less tail complex, which suggest a possible mechanism for DNA retention and ejection: a portal closed conformation temporarily retains the genome before the tail is assembled, whereas an open portal is found in the tail. Moreover, a fold including a seven-bladed β-propeller domain is described for the nozzle tail protein.
Organizational Affiliation:
Centro Nacional de Biotecnología, (CNB-CSIC), Darwin 3, 28049, Madrid, Spain.