Structure and Properties of a Natural Competence-Associated Pilin Suggest a Unique Pilus Tip-Associated DNA Receptor.
Salleh, M.Z., Karuppiah, V., Snee, M., Thistlethwaite, A., Levy, C.W., Knight, D., Derrick, J.P.(2019) mBio 10
- PubMed: 31186316
- DOI: https://doi.org/10.1128/mBio.00614-19
- Primary Citation of Related Structures:
6QVF, 6QVI - PubMed Abstract:
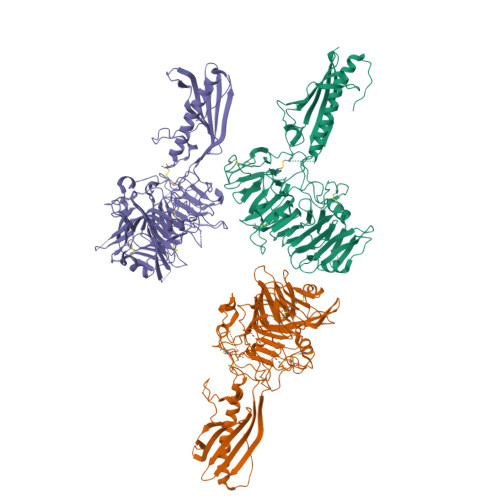
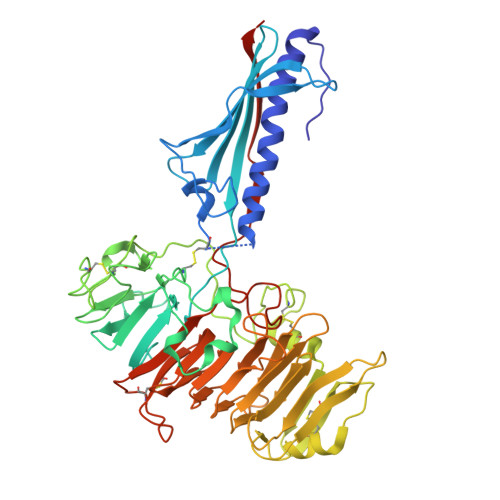
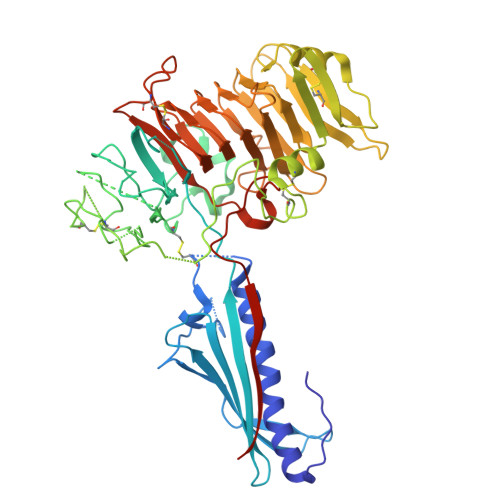
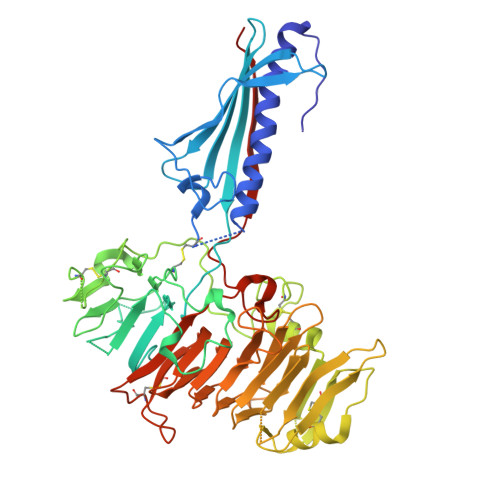
Natural competence is the term used to describe the uptake of "naked" extracellular DNA by bacteria; it plays a significant role in horizontal genetic exchange. It is associated with type IV pili, and specialized competence pili mediate DNA uptake. Here, we show that the crystal structure of a competence-associated protein from Thermus thermophilus , ComZ, consists of a type II secretion pseudopilin-like domain, with a large β-solenoid domain inserted into the β-sheet of the pilin-like fold. ComZ binds with high affinity to another competence-associated pilin, PilA2, which lies adjacent to the comZ gene in the genome. The crystal structure of PilA2 revealed a similar type II secretion pseudopilin-like fold, with a small subdomain; docking simulations predicted that PilA2 binds between the pseudopilin-like and β-solenoid domains of ComZ. Electrophoretic shift analysis and DNase protection studies were used to show that ComZ alone and the ComZ/PilA2 complex are able to bind DNA. Protection against reductive dimethylation was used in combination with mass spectrometry and site-directed mutagenesis to identify two lysine residues in ComZ which are involved in DNA binding. They are located between the two domains in ComZ, on the opposite side from the predicted PilA2 binding site. These results suggest a model in which PilA2 assists ComZ in forming the competence pilus tip and DNA binds to the side of the fiber. The results demonstrate how a type IV pilin can be adapted to a specific function by domain insertion and provide the first structural insights into a tip-located competence pilin. IMPORTANCE Thermus thermophilus is a thermophilic bacterium which is capable of natural transformation, the uptake of external DNA with high efficiency. DNA uptake is thought to be mediated by a competence-associated pilus, which binds the DNA substrate and mediates its transfer across the outer membrane and periplasm. Here, we describe the structural and functional analysis of two pilins which are known to be essential for DNA uptake, ComZ and PilA2. ComZ adopts an unusual structure, incorporating a large β-solenoid domain into the pilin structural framework. We argue on structural grounds that this structure cannot readily be accommodated into the competence pilus fiber unless it is at the tip. We also show that ComZ binds DNA and identify two lysine residues which appear to be important for DNA binding. These results suggest a model in which ComZ and PilA2 form a tip-associated DNA receptor which mediates DNA uptake.
Organizational Affiliation:
Lydia Becker Institute of Immunology and Inflammation, School of Biological Sciences, Faculty of Biology, Medicine and Health, Manchester Academic Health Science Centre, The University of Manchester, Manchester, United Kingdom.