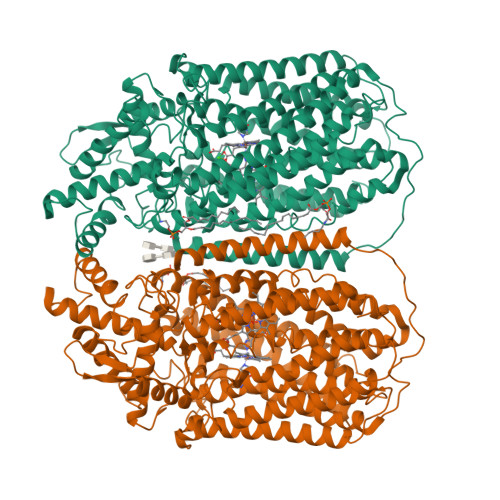
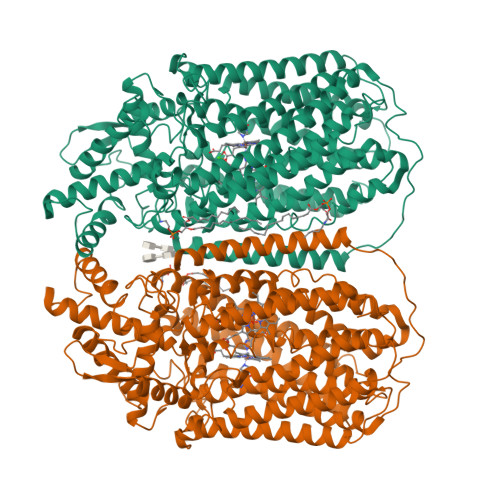
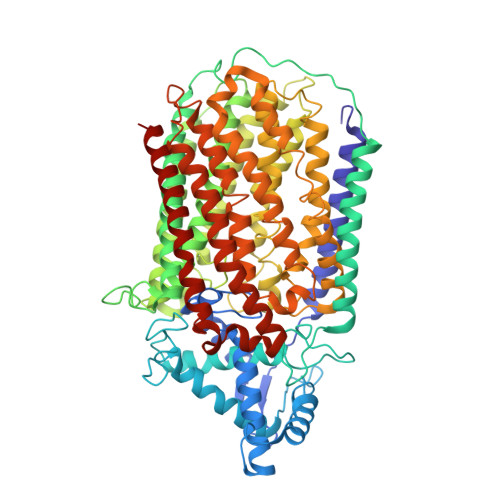
Dimeric structures of quinol-dependent nitric oxide reductases (qNORs) revealed by cryo-electron microscopy.
Gopalasingam, C.C., Johnson, R.M., Chiduza, G.N., Tosha, T., Yamamoto, M., Shiro, Y., Antonyuk, S.V., Muench, S.P., Hasnain, S.S.(2019) Sci Adv 5: eaax1803-eaax1803
- PubMed: 31489376
- DOI: https://doi.org/10.1126/sciadv.aax1803
- Primary Citation of Related Structures:
6QQ5, 6QQ6 - PubMed Abstract:
Quinol-dependent nitric oxide reductases (qNORs) are membrane-integrated, iron-containing enzymes of the denitrification pathway, which catalyze the reduction of nitric oxide (NO) to the major ozone destroying gas nitrous oxide (N 2 O). Cryo-electron microscopy structures of active qNOR from Alcaligenes xylosoxidans and an activity-enhancing mutant have been determined to be at local resolutions of 3.7 and 3.2 Å, respectively. They unexpectedly reveal a dimeric conformation (also confirmed for qNOR from Neisseria meningitidis ) and define the active-site configuration, with a clear water channel from the cytoplasm. Structure-based mutagenesis has identified key residues involved in proton transport and substrate delivery to the active site of qNORs. The proton supply direction differs from cytochrome c-dependent NOR (cNOR), where water molecules from the cytoplasm serve as a proton source similar to those from cytochrome c oxidase.
Organizational Affiliation:
Molecular Biophysics Group, Institute of Integrative Biology, Faculty of Health and Life Sciences, University of Liverpool, Liverpool L69 7ZB, UK.