Structure and ligand binding of the glutamine-II riboswitch.
Huang, L., Wang, J., Watkins, A.M., Das, R., Lilley, D.M.J.(2019) Nucleic Acids Res 47: 7666-7675
- PubMed: 31216023
- DOI: https://doi.org/10.1093/nar/gkz539
- Primary Citation of Related Structures:
6QN3 - PubMed Abstract:
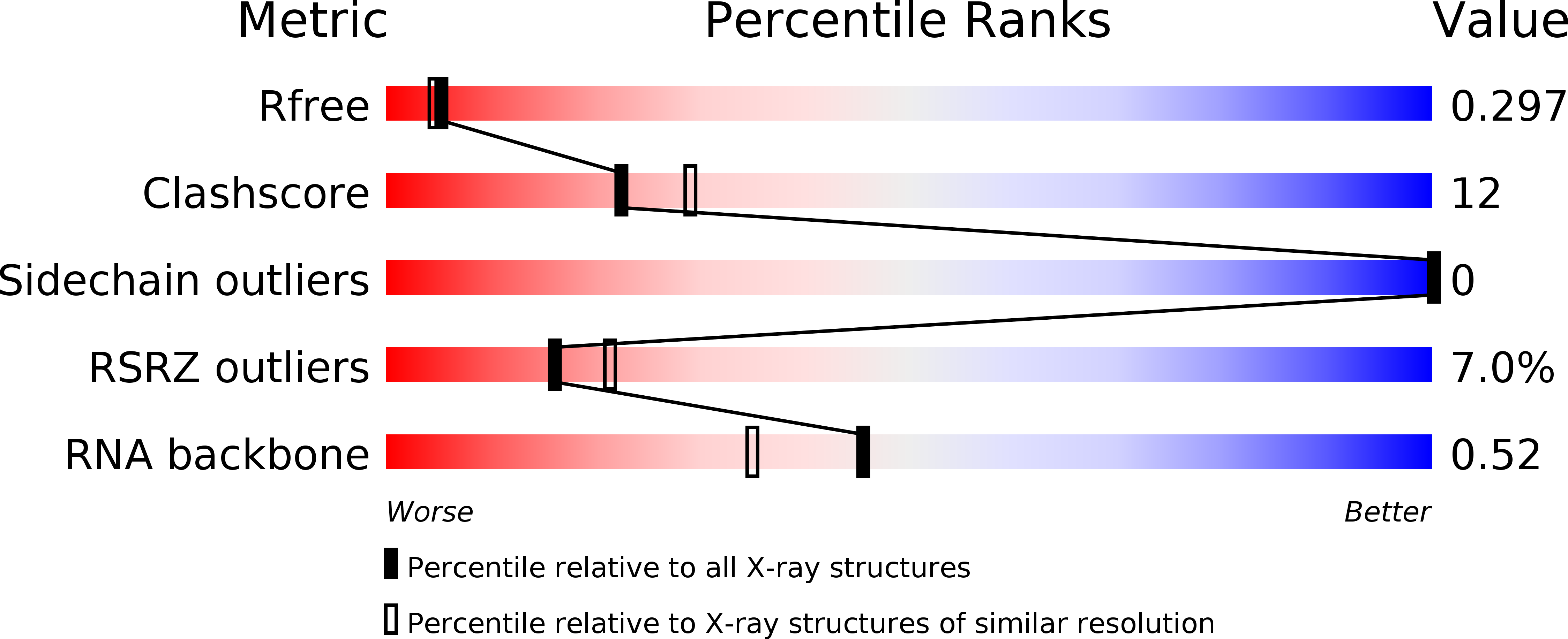
We have determined the structure of the glutamine-II riboswitch ligand binding domain using X-ray crystallography. The structure was solved using a novel combination of homology modeling and molecular replacement. The structure comprises three coaxial helical domains, the central one of which is a pseudoknot with partial triplex character. The major groove of this helix provides the binding site for L-glutamine, which is extensively hydrogen bonded to the RNA. Atomic mutation of the RNA at the ligand binding site leads to loss of binding shown by isothermal titration calorimetry, explaining the specificity of the riboswitch. A metal ion also plays an important role in ligand binding. This is directly bonded to a glutamine carboxylate oxygen atom, and its remaining inner-sphere water molecules make hydrogen bonding interactions with the RNA.
Organizational Affiliation:
Cancer Research UK Nucleic Acid Structure Research Group, MSI/WTB Complex, The University of Dundee, Dow Street, Dundee DD1 5EH, UK.