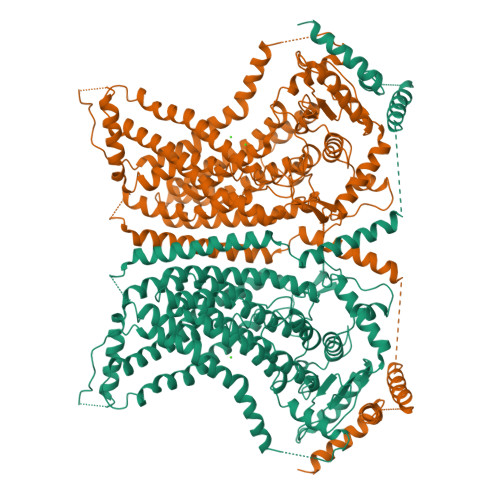
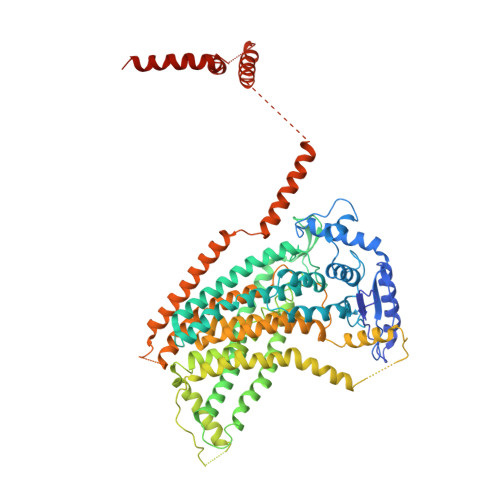
Stepwise activation mechanism of the scramblase nhTMEM16 revealed by cryo-EM.
Kalienkova, V., Clerico Mosina, V., Bryner, L., Oostergetel, G.T., Dutzler, R., Paulino, C.(2019) Elife 8
- PubMed: 30785398
- DOI: https://doi.org/10.7554/eLife.44364
- Primary Citation of Related Structures:
6QM4, 6QM5, 6QM6, 6QM9, 6QMA, 6QMB - PubMed Abstract:
Scramblases catalyze the movement of lipids between both leaflets of a bilayer. Whereas the X-ray structure of the protein nhTMEM16 has previously revealed the architecture of a Ca 2+ -dependent lipid scramblase, its regulation mechanism has remained elusive. Here, we have used cryo-electron microscopy and functional assays to address this question. Ca 2+ -bound and Ca 2+ -free conformations of nhTMEM16 in detergent and lipid nanodiscs illustrate the interactions with its environment and they reveal the conformational changes underlying its activation. In this process, Ca 2+ binding induces a stepwise transition of the catalytic subunit cavity, converting a closed cavity that is shielded from the membrane in the absence of ligand, into a polar furrow that becomes accessible to lipid headgroups in the Ca 2+ -bound state. Additionally, our structures demonstrate how nhTMEM16 distorts the membrane at both entrances of the subunit cavity, thereby decreasing the energy barrier for lipid movement.
Organizational Affiliation:
Department of Biochemistry, University of Zurich, Zurich, Switzerland.