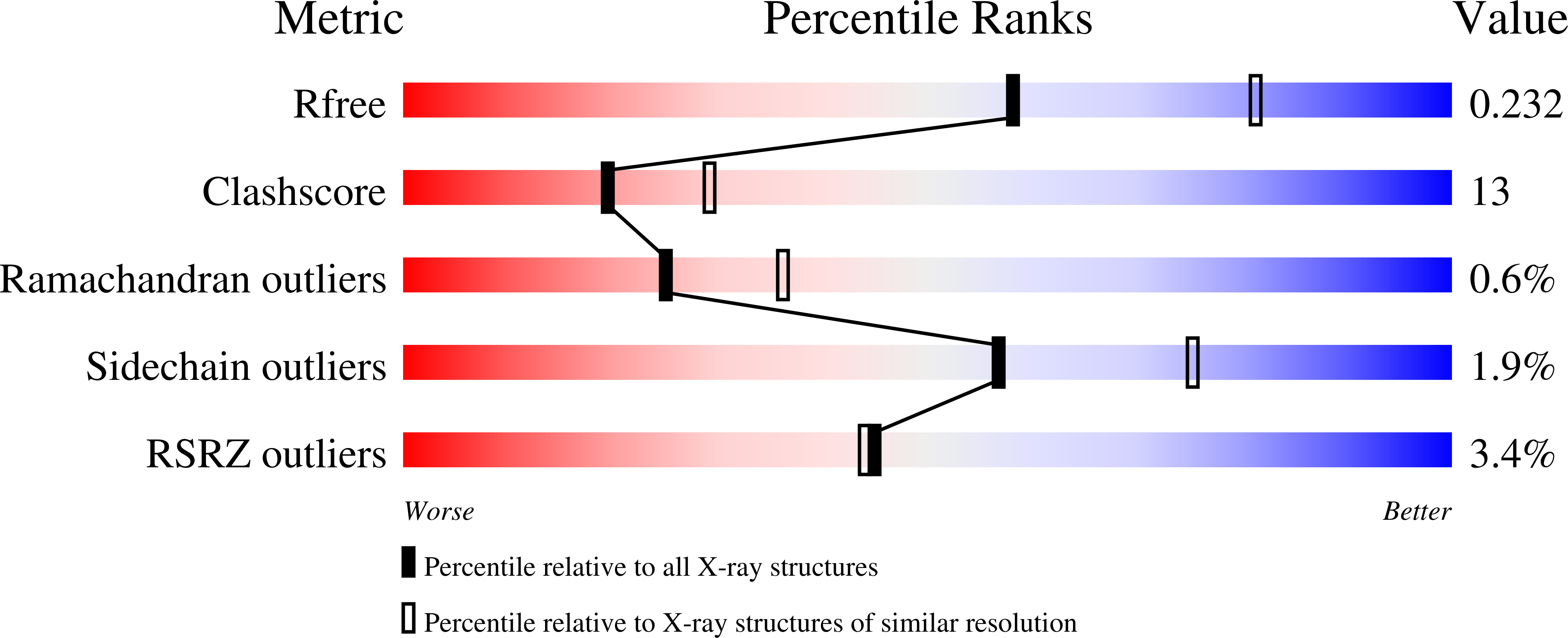
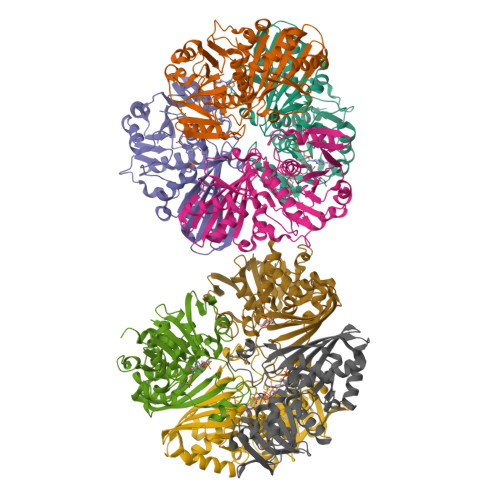
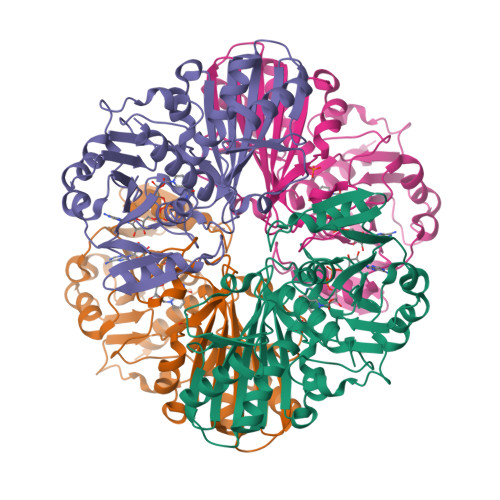
Thermal stability and structure of glyceraldehyde-3-phosphate dehydrogenase from the coral Acropora millepora.
Perez, A.M., Wolfe, J.A., Schermerhorn, J.T., Qian, Y., Cela, B.A., Kalinowski, C.R., Largoza, G.E., Fields, P.A., Brandt, G.S.(2021) RSC Adv 11: 10364-10374
- PubMed: 35423531
- DOI: https://doi.org/10.1039/d0ra10119b
- Primary Citation of Related Structures:
6PX2 - PubMed Abstract:
Corals are vulnerable to increasing ocean temperatures. It is known that elevated temperatures lead to the breakdown of an essential mutualistic relationship with photosynthetic algae. The molecular mechanisms of this temperature-dependent loss of symbiosis are less well understood. Here, the thermal stability of a critical metabolic enzyme, glyceraldehyde-3-phosphate dehydrogenase, from the stony coral Acropora millepora was found to increase significantly in the presence of its cofactor NAD + . Determination of the structure of the cofactor-enzyme complex (PDB ID 6PX2) revealed variable NAD + occupancy across the four monomers of the tetrameric enzyme. The structure of the fully occupied monomers was compared to those with partial cofactor occupancy, identifying regions of difference that may account for the increased thermal stability.
Organizational Affiliation:
Department of Chemistry, Franklin & Marshall College Lancaster PA 17604 USA gbrandt@fandm.edu +1 717 358 4846 +1 717 358 4846.