Biophysical and X-ray structural studies of the (GGGTT)3GGG G-quadruplex in complex with N-methyl mesoporphyrin IX.
Lin, L.Y., McCarthy, S., Powell, B.M., Manurung, Y., Xiang, I.M., Dean, W.L., Chaires, B., Yatsunyk, L.A.(2020) PLoS One 15: e0241513-e0241513
- PubMed: 33206666
- DOI: https://doi.org/10.1371/journal.pone.0241513
- Primary Citation of Related Structures:
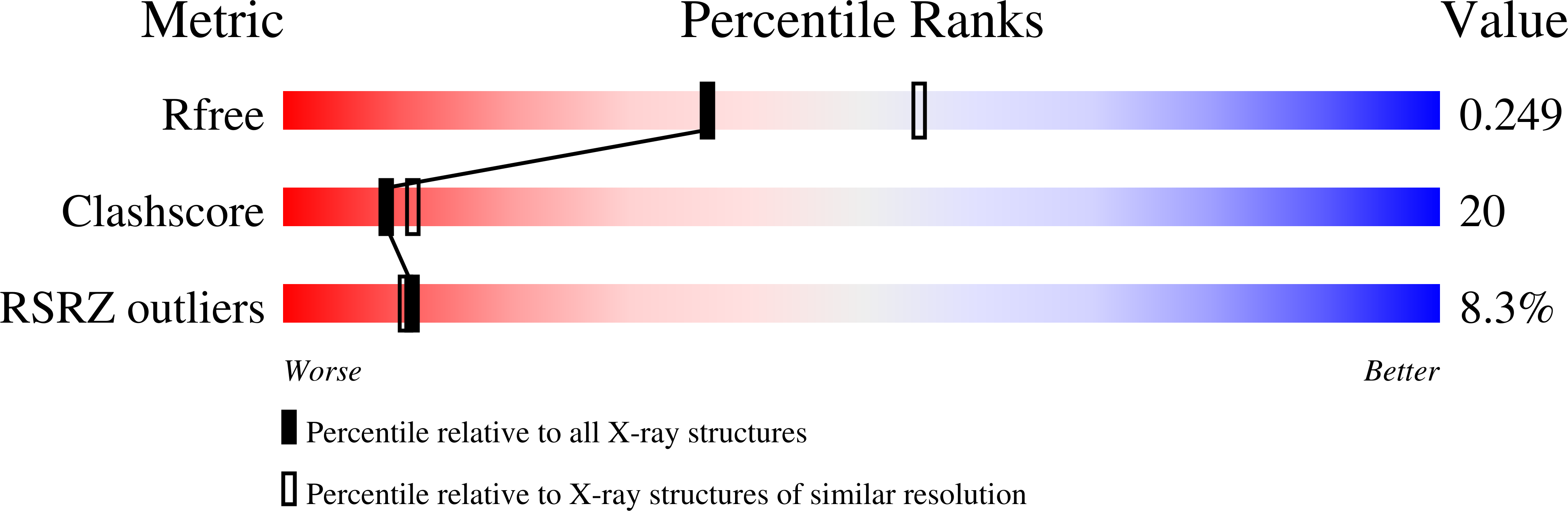
6P45, 6PNK - PubMed Abstract:
The G-quadruplex (GQ) is a well-studied non-canonical DNA structure formed by G-rich sequences found at telomeres and gene promoters. Biological studies suggest that GQs may play roles in regulating gene expression, DNA replication, and DNA repair. Small molecule ligands were shown to alter GQ structure and stability and thereby serve as novel therapies, particularly against cancer. In this work, we investigate the interaction of a G-rich sequence, 5'-GGGTTGGGTTGGGTTGGG-3' (T1), with a water-soluble porphyrin, N-methyl mesoporphyrin IX (NMM) via biophysical and X-ray crystallographic studies. UV-vis and fluorescence titrations, as well as a Job plot, revealed a 1:1 binding stoichiometry with an impressively tight binding constant of 30-50 μM-1 and ΔG298 of -10.3 kcal/mol. Eight extended variants of T1 (named T2 -T9) were fully characterized and T7 was identified as a suitable candidate for crystallographic studies. We solved the crystal structures of the T1- and T7-NMM complexes at 2.39 and 2.34 Å resolution, respectively. Both complexes form a 5'-5' dimer of parallel GQs capped by NMM at the 3' G-quartet, supporting the 1:1 binding stoichiometry. Our work provides invaluable details about GQ-ligand binding interactions and informs the design of novel anticancer drugs that selectively recognize specific GQs and modulate their stability for therapeutic purposes.
Organizational Affiliation:
Department of Chemistry and Biochemistry, Swarthmore College, Swarthmore, Pennsylvania, United States of America.