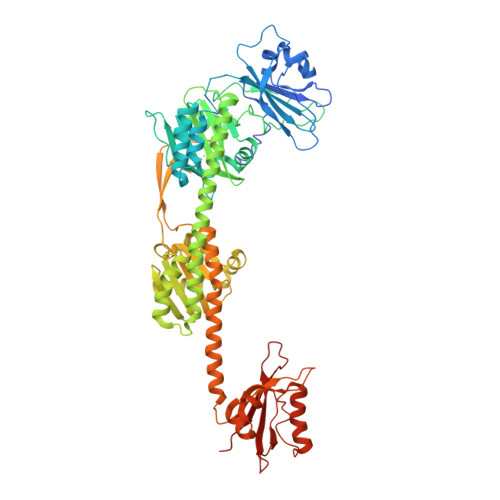
Structural basis for the Pr-Pfr long-range signaling mechanism of a full-length bacterial phytochrome at the atomic level.
Otero, L.H., Foscaldi, S., Antelo, G.T., Rosano, G.L., Sirigu, S., Klinke, S., Defelipe, L.A., Sanchez-Lamas, M., Battocchio, G., Conforte, V., Vojnov, A.A., Chavas, L.M.G., Goldbaum, F.A., Mroginski, M.A., Rinaldi, J., Bonomi, H.R.(2021) Sci Adv 7: eabh1097-eabh1097
- PubMed: 34818032
- DOI: https://doi.org/10.1126/sciadv.abh1097
- Primary Citation of Related Structures:
6PL0, 7L59, 7L5A - PubMed Abstract:
Phytochromes constitute a widespread photoreceptor family that typically interconverts between two photostates called Pr (red light–absorbing) and Pfr (far-red light–absorbing). The lack of full-length structures solved at the (near-)atomic level in both pure Pr and Pfr states leaves gaps in the structural mechanisms involved in the signal transmission pathways during the photoconversion. Here, we present the crystallographic structures of three versions from the plant pathogen Xanthomonas campestris virulence regulator Xcc BphP bacteriophytochrome, including two full-length proteins, in the Pr and Pfr states. The structures show a reorganization of the interaction networks within and around the chromophore-binding pocket, an α-helix/β-sheet tongue transition, and specific domain reorientations, along with interchanging kinks and breaks at the helical spine as a result of the photoswitching, which subsequently affect the quaternary assembly. These structural findings, combined with multidisciplinary studies, allow us to describe the signaling mechanism of a full-length bacterial phytochrome at the atomic level.
Organizational Affiliation:
Fundación Instituto Leloir, IIBBA-CONICET, Av. Patricias Argentinas 435 (C1405BWE), Buenos Aires, Argentina.