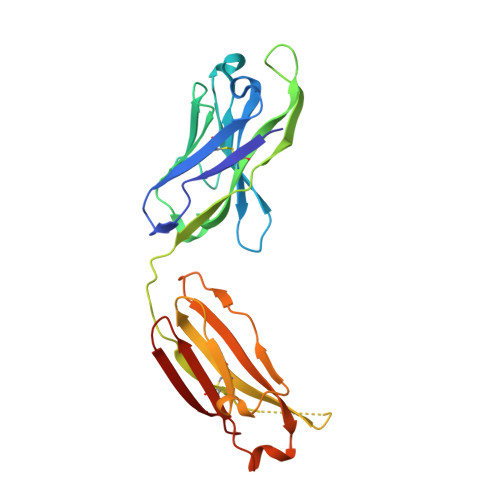
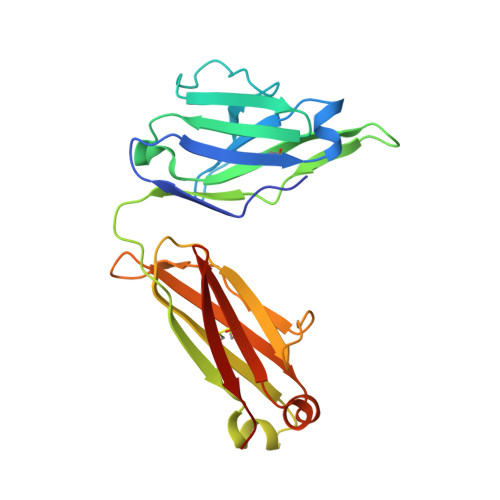
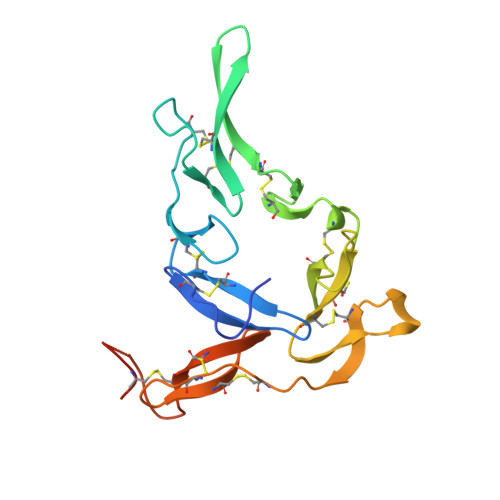
Potent antibody lineage against malaria transmission elicited by human vaccination with Pfs25.
McLeod, B., Miura, K., Scally, S.W., Bosch, A., Nguyen, N., Shin, H., Kim, D., Volkmuth, W., Ramisch, S., Chichester, J.A., Streatfield, S., Woods, C., Schief, W.R., Emerling, D., King, C.R., Julien, J.P.(2019) Nat Commun 10: 4328-4328
- PubMed: 31551421
- DOI: https://doi.org/10.1038/s41467-019-11980-6
- Primary Citation of Related Structures:
6PHB, 6PHC, 6PHD, 6PHF, 6PHG, 6PHH - PubMed Abstract:
Transmission-blocking vaccines have the potential to be key contributors to malaria elimination. Such vaccines elicit antibodies that inhibit parasites during their development in Anopheles mosquitoes, thus breaking the cycle of transmission. To date, characterization of humoral responses to Plasmodium falciparum transmission-blocking vaccine candidate Pfs25 has largely been conducted in pre-clinical models. Here, we present molecular analyses of human antibody responses generated in a clinical trial evaluating Pfs25 vaccination. From a collection of monoclonal antibodies with transmission-blocking activity, we identify the most potent transmission-blocking antibody yet described against Pfs25; 2544. The interactions of 2544 and three other antibodies with Pfs25 are analyzed by crystallography to understand structural requirements for elicitation of human transmission-blocking responses. Our analyses provide insights into Pfs25 immunogenicity and epitope potency, and detail an affinity maturation pathway for a potent transmission-blocking antibody in humans. Our findings can be employed to guide the design of improved malaria transmission-blocking vaccines.
Organizational Affiliation:
Program in Molecular Medicine, The Hospital for Sick Children Research Institute, 686 Bay Street, Toronto, ON, M5G 0A4, Canada.