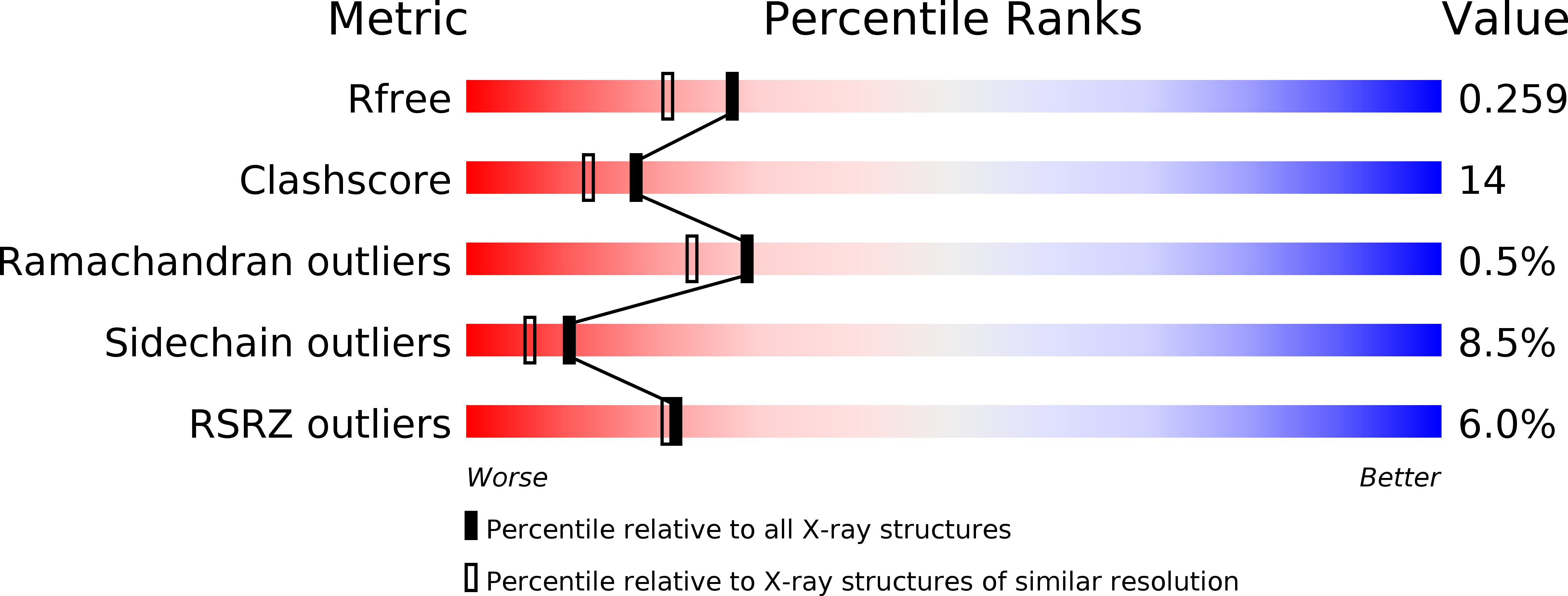
The SrrAB two-component system regulatesStaphylococcus aureuspathogenicity through redox sensitive cysteines.
Tiwari, N., Lopez-Redondo, M., Miguel-Romero, L., Kulhankova, K., Cahill, M.P., Tran, P.M., Kinney, K.J., Kilgore, S.H., Al-Tameemi, H., Herfst, C.A., Tuffs, S.W., Kirby, J.R., Boyd, J.M., McCormick, J.K., Salgado-Pabon, W., Marina, A., Schlievert, P.M., Fuentes, E.J.(2020) Proc Natl Acad Sci U S A 117: 10989-10999
- PubMed: 32354997
- DOI: https://doi.org/10.1073/pnas.1921307117
- Primary Citation of Related Structures:
6PAJ - PubMed Abstract:
Staphylococcus aureus infections can lead to diseases that range from localized skin abscess to life-threatening toxic shock syndrome. The SrrAB two-component system (TCS) is a global regulator of S. aureus virulence and critical for survival under environmental conditions such as hypoxic, oxidative, and nitrosative stress found at sites of infection. Despite the critical role of SrrAB in S. aureus pathogenicity, the mechanism by which the SrrAB TCS senses and responds to these environmental signals remains unknown. Bioinformatics analysis showed that the SrrB histidine kinase contains several domains, including an extracellular Cache domain and a cytoplasmic HAMP-PAS-DHp-CA region. Here, we show that the PAS domain regulates both kinase and phosphatase enzyme activity of SrrB and present the structure of the DHp-CA catalytic core. Importantly, this structure shows a unique intramolecular cysteine disulfide bond in the ATP-binding domain that significantly affects autophosphorylation kinetics. In vitro data show that the redox state of the disulfide bond affects S. aureus biofilm formation and toxic shock syndrome toxin-1 production. Moreover, with the use of the rabbit infective endocarditis model, we demonstrate that the disulfide bond is a critical regulatory element of SrrB function during S. aureus infection. Our data support a model whereby the disulfide bond and PAS domain of SrrB sense and respond to the cellular redox environment to regulate S. aureus survival and pathogenesis.
Organizational Affiliation:
Department of Biochemistry, University of Iowa, Iowa City, IA 52242.