Salmonella Typhoid Toxin PltB Subunit and Its Non-typhoidal Salmonella Ortholog Confer Differential Host Adaptation and Virulence.
Lee, S., Yang, Y.A., Milano, S.K., Nguyen, T., Ahn, C., Sim, J.H., Thompson, A.J., Hillpot, E.C., Yoo, G., Paulson, J.C., Song, J.(2020) Cell Host Microbe 27: 937-949.e6
- PubMed: 32396840
- DOI: https://doi.org/10.1016/j.chom.2020.04.005
- Primary Citation of Related Structures:
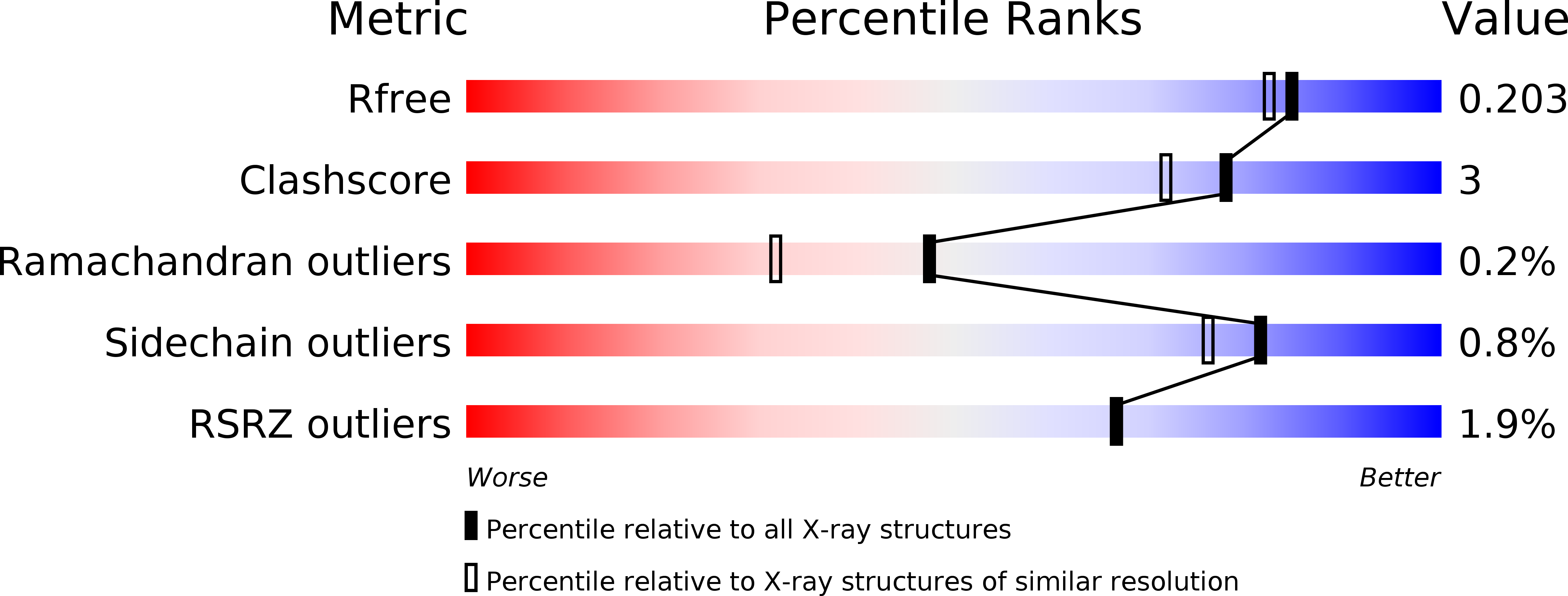
6P4M, 6P4N, 6P4P, 6P4Q, 6P4R, 6P4S, 6P4T - PubMed Abstract:
Typhoidal and non-typhoidal Salmonelleae (NTS) cause typhoid fever and gastroenteritis, respectively, in humans. Salmonella typhoid toxin contributes to typhoid disease progression and chronic infection, but little is known about the role of its NTS ortholog. We found that typhoid toxin and its NTS ortholog induce different clinical presentations. The PltB subunit of each toxin exhibits different glycan-binding preferences that correlate with glycan expression profiles of host cells targeted by each bacterium at the primary infection or intoxication sites. Through co-crystal structures of PltB subunits bound to specific glycan receptor moieties, we show that they induce markedly different glycan-binding preferences and virulence outcomes. Furthermore, immunization with the NTS S. Javiana or its toxin offers cross-reactive protection against lethal-dose typhoid toxin challenge. Cumulatively, these results offer insights into the evolution of host adaptations in Salmonella AB toxins, their cell and tissue tropisms, and the design for improved typhoid vaccines and therapeutics.
Organizational Affiliation:
Department of Microbiology and Immunology, Cornell University College of Veterinary Medicine, Ithaca, NY 14853, USA.