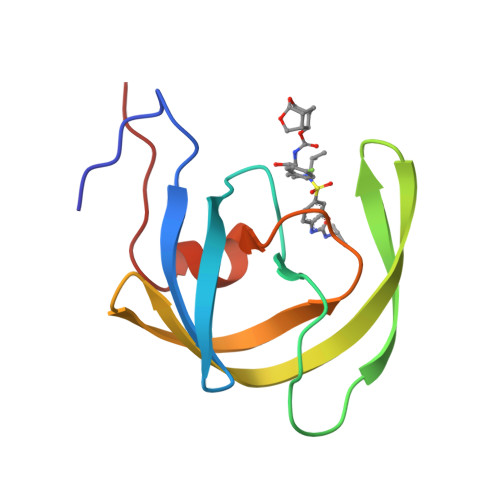
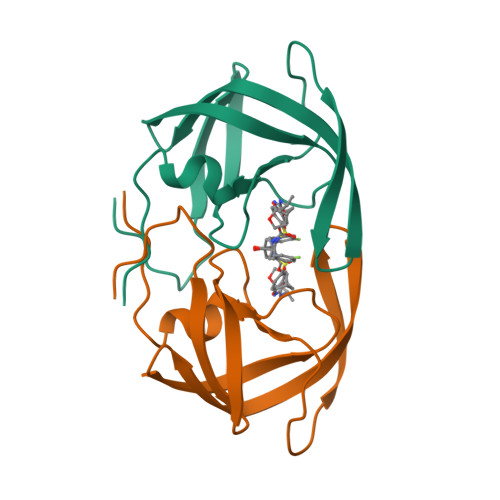
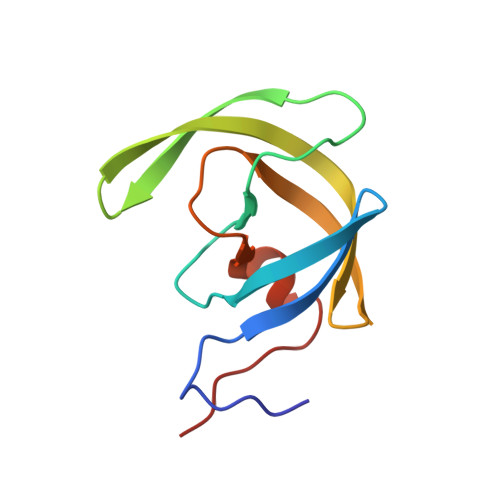
Single atom changes in newly synthesized HIV protease inhibitors reveal structural basis for extreme affinity, high genetic barrier, and adaptation to the HIV protease plasticity.
Bulut, H., Hattori, S.I., Aoki-Ogata, H., Hayashi, H., Das, D., Aoki, M., Davis, D.A., Rao, K.V., Nyalapatla, P.R., Ghosh, A.K., Mitsuya, H.(2020) Sci Rep 10: 10664-10664
- PubMed: 32606378
- DOI: https://doi.org/10.1038/s41598-020-65993-z
- Primary Citation of Related Structures:
6OGL, 6OGP, 6OGQ, 6OGS, 6OGT, 6OYD, 6OYR - PubMed Abstract:
HIV-1 protease inhibitors (PIs), such as darunavir (DRV), are the key component of antiretroviral therapy. However, HIV-1 often acquires resistance to PIs. Here, seven novel PIs were synthesized, by introducing single atom changes such as an exchange of a sulfur to an oxygen, scission of a single bond in P2'-cyclopropylaminobenzothiazole (or -oxazole), and/or P1-benzene ring with fluorine scan of mono- or bis-fluorine atoms around DRV's scaffold. X-ray structural analyses of the PIs complexed with wild-type Protease (PR WT ) and highly-multi-PI-resistance-associated PR DRV R P51 revealed that the PIs better adapt to structural plasticity in PR with resistance-associated amino acid substitutions by formation of optimal sulfur bond and adaptation of cyclopropyl ring in the S2'-subsite. Furthermore, these PIs displayed increased cell permeability and extreme anti-HIV-1 potency compared to DRV. Our work provides the basis for developing novel PIs with high potency against PI-resistant HIV-1 variants with a high genetic barrier.
Organizational Affiliation:
HIV and AIDS Malignancy Branch, National Cancer Institute, National Institutes of Health, Bethesda, 20892, MD, United States.