Casposase structure and the mechanistic link between DNA transposition and spacer acquisition by CRISPR-Cas.
Hickman, A.B., Kailasan, S., Genzor, P., Haase, A.D., Dyda, F.(2020) Elife 9
- PubMed: 31913120
- DOI: https://doi.org/10.7554/eLife.50004
- Primary Citation of Related Structures:
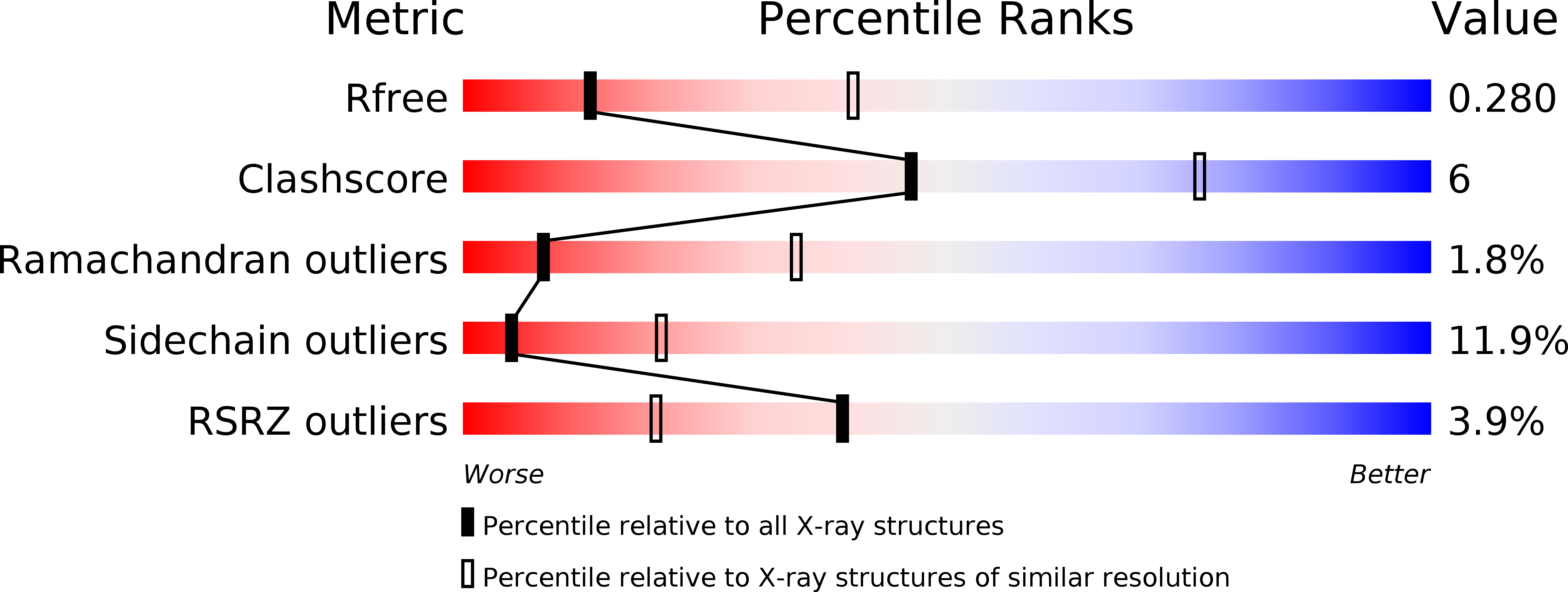
6OPM - PubMed Abstract:
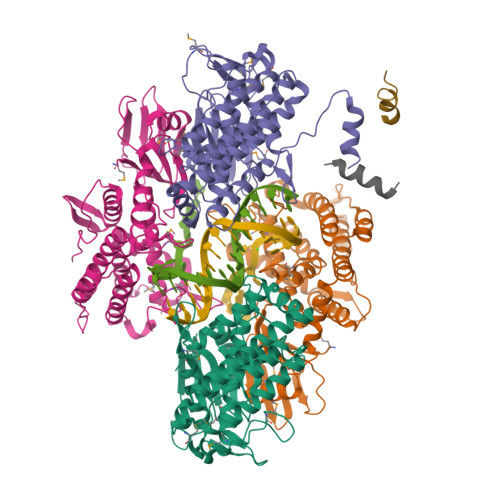
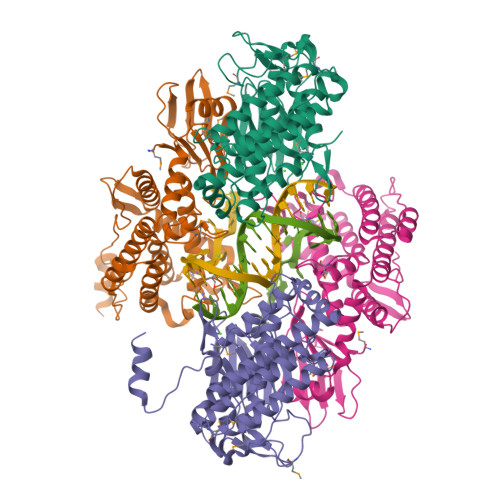
Key to CRISPR-Cas adaptive immunity is maintaining an ongoing record of invading nucleic acids, a process carried out by the Cas1-Cas2 complex that integrates short segments of foreign genetic material (spacers) into the CRISPR locus. It is hypothesized that Cas1 evolved from casposases, a novel class of transposases. We show here that the Methanosarcina mazei casposase can integrate varied forms of the casposon end in vitro, and recapitulates several properties of CRISPR-Cas integrases including site-specificity. The X-ray structure of the casposase bound to DNA representing the product of integration reveals a tetramer with target DNA bound snugly between two dimers in which single-stranded casposon end binding resembles that of spacer 3'-overhangs. The differences between transposase and CRISPR-Cas integrase are largely architectural, and it appears that evolutionary change involved changes in protein-protein interactions to favor Cas2 binding over tetramerization; this in turn led to preferred integration of single spacers over two transposon ends.
Organizational Affiliation:
Laboratory of Molecular Biology, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, United States.