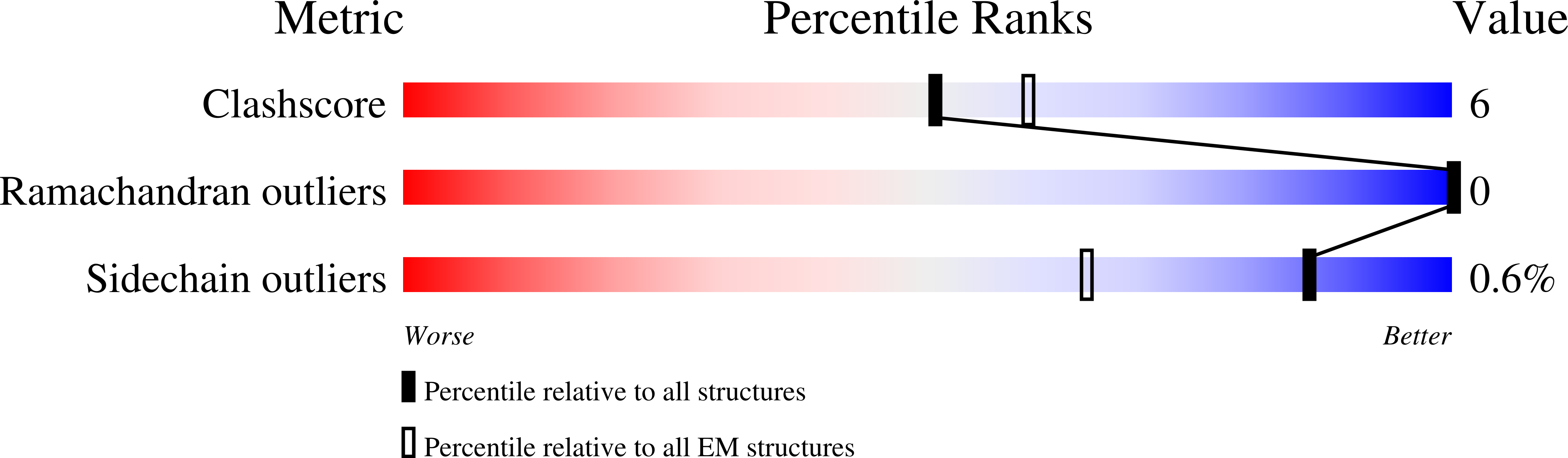CryoEM structures of Arabidopsis DDR complexes involved in RNA-directed DNA methylation.
Wongpalee, S.P., Liu, S., Gallego-Bartolome, J., Leitner, A., Aebersold, R., Liu, W., Yen, L., Nohales, M.A., Kuo, P.H., Vashisht, A.A., Wohlschlegel, J.A., Feng, S., Kay, S.A., Zhou, Z.H., Jacobsen, S.E.(2019) Nat Commun 10: 3916-3916
- PubMed: 31477705
- DOI: https://doi.org/10.1038/s41467-019-11759-9
- Primary Citation of Related Structures:
6OIS, 6OIT - PubMed Abstract:
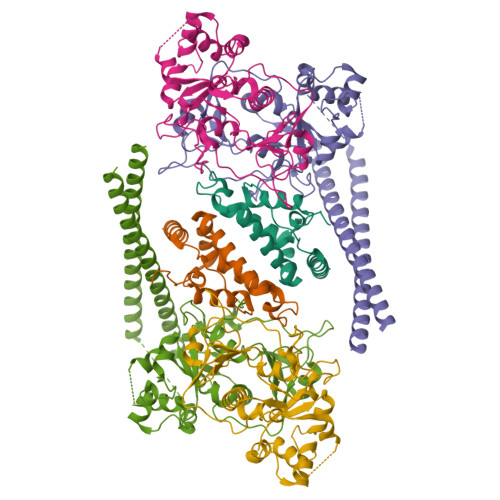
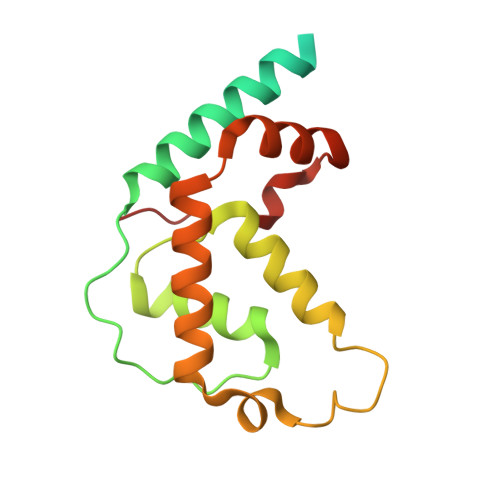
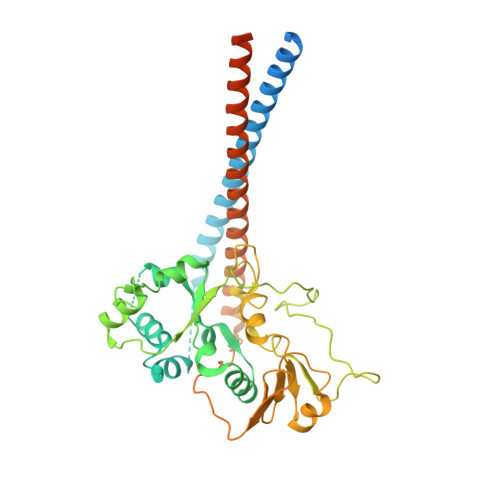
Transcription by RNA polymerase V (Pol V) in plants is required for RNA-directed DNA methylation, leading to transcriptional gene silencing. Global chromatin association of Pol V requires components of the DDR complex DRD1, DMS3 and RDM1, but the assembly process of this complex and the underlying mechanism for Pol V recruitment remain unknown. Here we show that all DDR complex components co-localize with Pol V, and we report the cryoEM structures of two complexes associated with Pol V recruitment-DR (DMS3-RDM1) and DDR' (DMS3-RDM1-DRD1 peptide), at 3.6 Å and 3.5 Å resolution, respectively. RDM1 dimerization at the center frames the assembly of the entire complex and mediates interactions between DMS3 and DRD1 with a stoichiometry of 1 DRD1:4 DMS3:2 RDM1. DRD1 binding to the DR complex induces a drastic movement of a DMS3 coiled-coil helix bundle. We hypothesize that both complexes are functional intermediates that mediate Pol V recruitment.
Organizational Affiliation:
Department of Molecular, Cellular and Developmental Biology, University of California, Los Angeles (UCLA), Los Angeles, CA, 90095, USA.