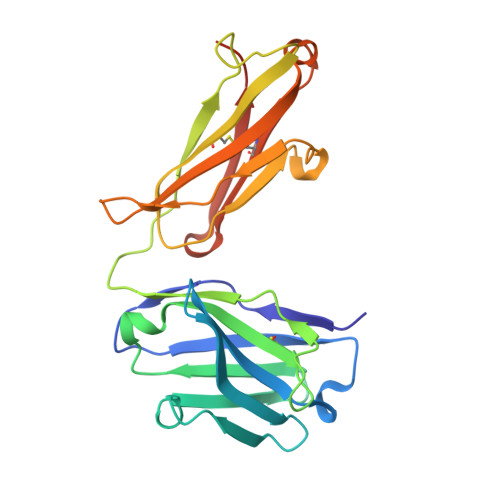
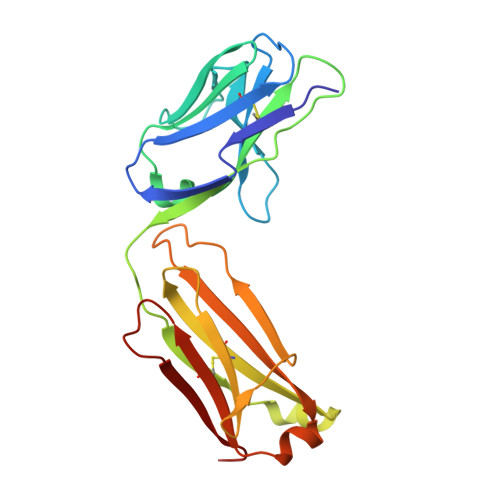
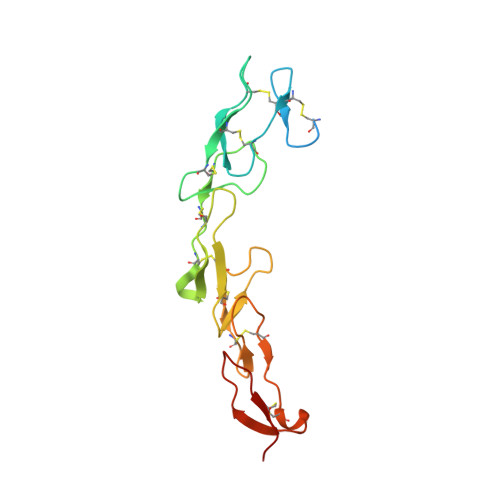
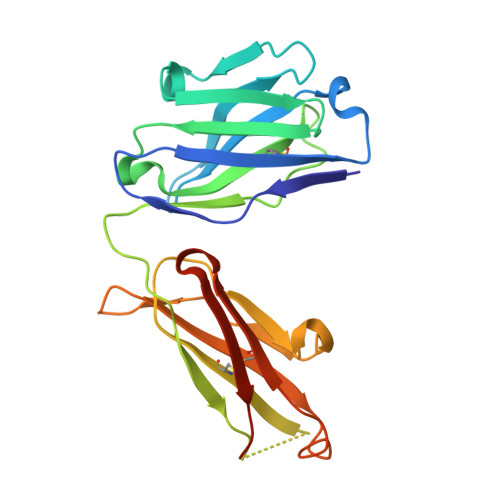
Tetravalent biepitopic targeting enables intrinsic antibody agonism of tumor necrosis factor receptor superfamily members.
Yang, Y., Yeh, S.H., Madireddi, S., Matochko, W.L., Gu, C., Pacheco Sanchez, P., Ultsch, M., De Leon Boenig, G., Harris, S.F., Leonard, B., Scales, S.J., Zhu, J.W., Christensen, E., Hang, J.Q., Brezski, R.J., Marsters, S., Ashkenazi, A., Sukumaran, S., Chiu, H., Cubas, R., Kim, J.M., Lazar, G.A.(2019) MAbs 11: 996-1011
- PubMed: 31156033
- DOI: https://doi.org/10.1080/19420862.2019.1625662
- Primary Citation of Related Structures:
6OGX, 6OKM, 6OKN - PubMed Abstract:
Agonism of members of the tumor necrosis factor receptor superfamily (TNFRSF) with monoclonal antibodies is of high therapeutic interest due to their role in immune regulation and cell proliferation. A major hurdle for pharmacologic activation of this receptor class is the requirement for high-order clustering, a mechanism that imposes a reliance in vivo on Fc receptor-mediated crosslinking. This extrinsic dependence represents a potential limitation of virtually the entire pipeline of agonist TNFRSF antibody drugs, of which none have thus far been approved or reached late-stage clinical trials. We show that tetravalent biepitopic targeting enables robust intrinsic antibody agonism for two members of this family, OX40 and DR5, that is superior to extrinsically crosslinked native parental antibodies. Tetravalent biepitopic anti-OX40 engagement co-stimulated OX40 low cells, obviated the requirement for CD28 co-signal for T cell activation, and enabled superior pharmacodynamic activity relative to native IgG in a murine vaccination model. This work establishes a proof of concept for an engineering approach that addresses a major gap for the therapeutic activation of this important receptor class.
Organizational Affiliation:
a Departments of Antibody Engineering, Genentech Inc ., South San Francisco , CA , USA.