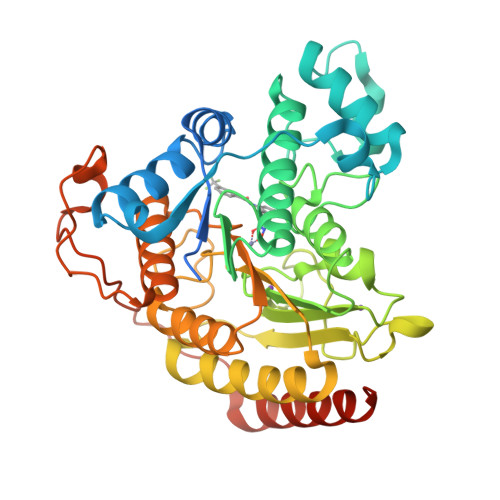
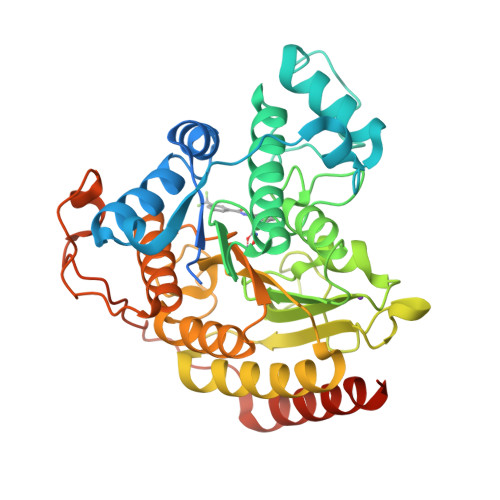
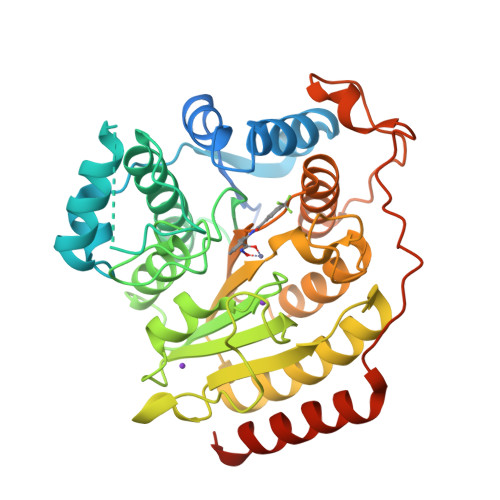
Structure-based Discovery of Novel N-(E)-N-Hydroxy-3-(2-(2-oxoimidazolidin-1-yl)phenyl)acrylamides as Potent and Selective HDAC8 inhibitors
Zheng, X., Conti, C., Caravella, J., Zablocki, M.-M., Bair, K., Barczak, N., Han, B., Lancia Jr., D., Liu, C., Martin, M., Ng, P.Y., Rudnitskaya, A., Thomason, J.J., Garcia-Dancey, R., Hardy, C., Lahdenranta, J., Leng, C., Li, P., Pardo, E., Saldahna, A., Tan, T., Toms, A.V., Yao, L., Zhang, C.To be published.