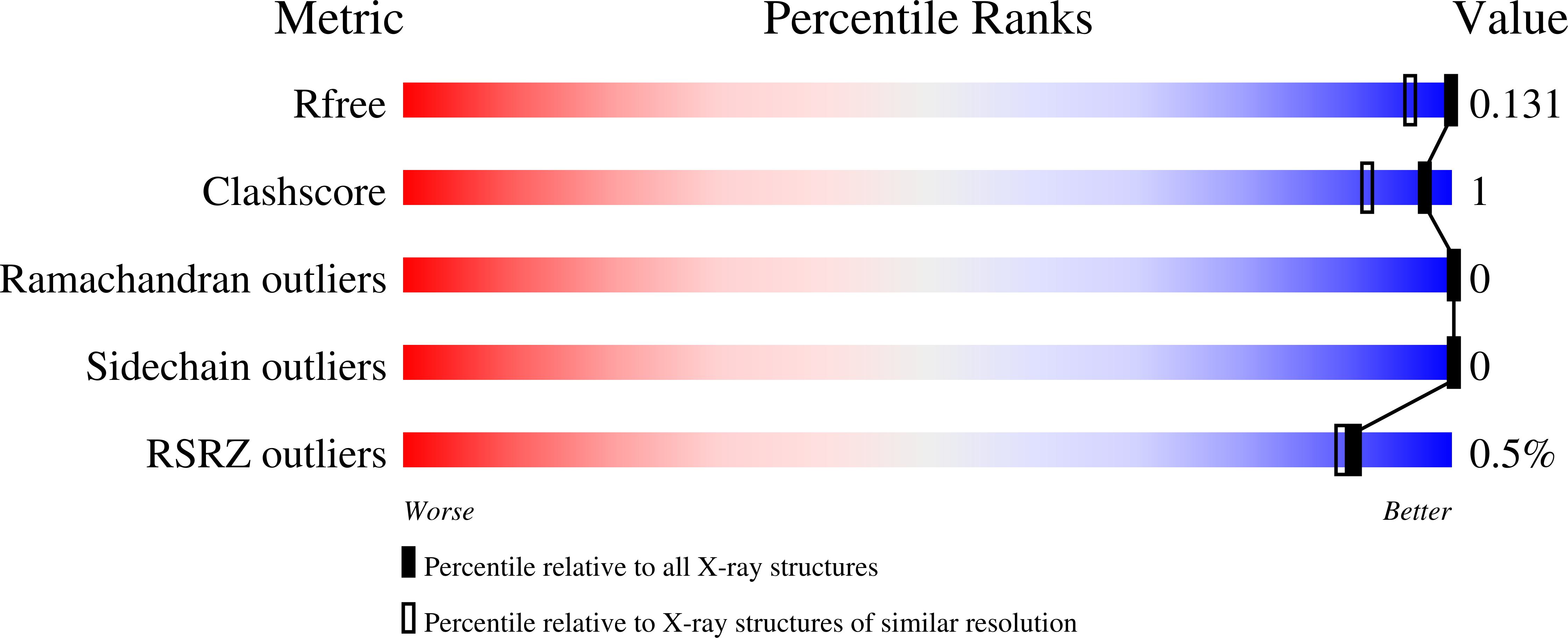
Substitutions of Amino Acid Residues in the Substrate Binding Site of Horse Liver Alcohol Dehydrogenase Have Small Effects on the Structures but Significantly Affect Catalysis of Hydrogen Transfer.
Kim, K., Plapp, B.V.(2020) Biochemistry 59: 862-879
- PubMed: 31994873
- DOI: https://doi.org/10.1021/acs.biochem.9b01074
- Primary Citation of Related Structures:
6O91, 6OA7, 6OWM, 6OWP - PubMed Abstract:
Previous studies showed that the L57F and F93W alcohol dehydrogenases catalyze the oxidation of benzyl alcohol with some quantum mechanical hydrogen tunneling, whereas the V203A enzyme has diminished tunneling. Here, steady-state kinetics for the L57F and F93W enzymes were studied, and microscopic rate constants for the ordered bi-bi mechanism were estimated from simulations of transient kinetics for the S48T, F93A, S48T/F93A, F93W, and L57F enzymes. Catalytic efficiencies for benzyl alcohol oxidation ( V 1 / E t K b ) vary over a range of ∼100-fold for the less active enzymes up to the L57F enzyme and are mostly associated with the binding of alcohol rather than the rate constants for hydride transfer. In contrast, catalytic efficiencies for benzaldehyde reduction ( V 2 / E t K p ) are ∼500-fold higher for the L57F enzyme than for the less active enzymes and are mostly associated with the rate constants for hydride transfer. Atomic-resolution structures (1.1 Å) for the F93W and L57F enzymes complexed with NAD + and 2,3,4,5,6-pentafluorobenzyl alcohol or 2,2,2-trifluoroethanol are almost identical to previous structures for the wild-type, S48T, and V203A enzymes. Least-squares refinement with SHELXL shows that the nicotinamide ring is slightly strained in all complexes and that the apparent donor-acceptor distances from C4N of NAD to C7 of pentafluorobenzyl alcohol range from 3.28 to 3.49 Å (±0.02 Å) and are not correlated with the rate constants for hydride transfer or hydrogen tunneling. How the substitutions affect the dynamics of reorganization during hydrogen transfer and the extent of tunneling remain to be determined.
Organizational Affiliation:
Department of Biochemistry , The University of Iowa , Iowa City , Iowa 52242 , United States.