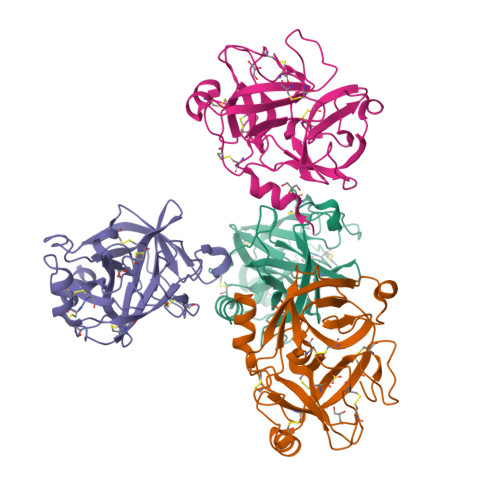
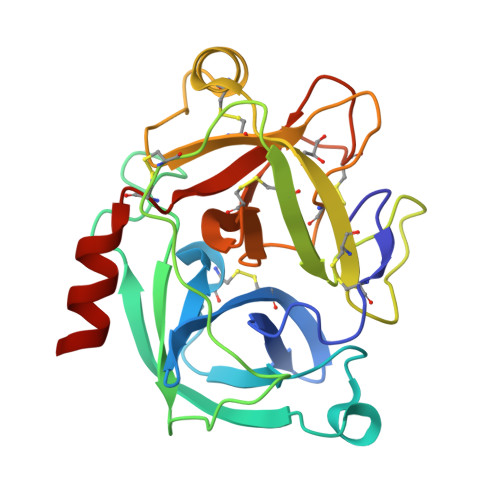
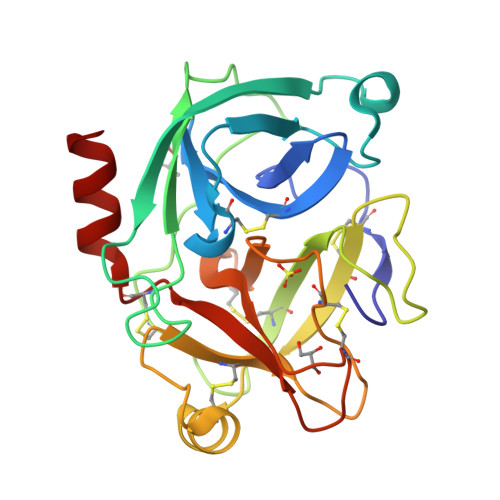
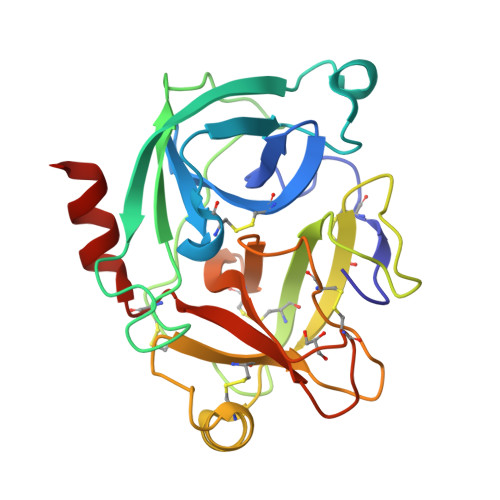
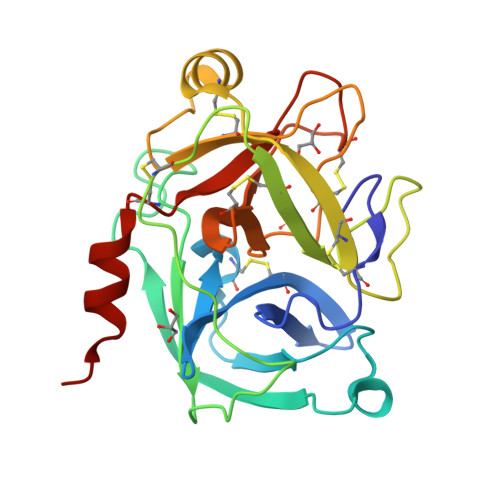
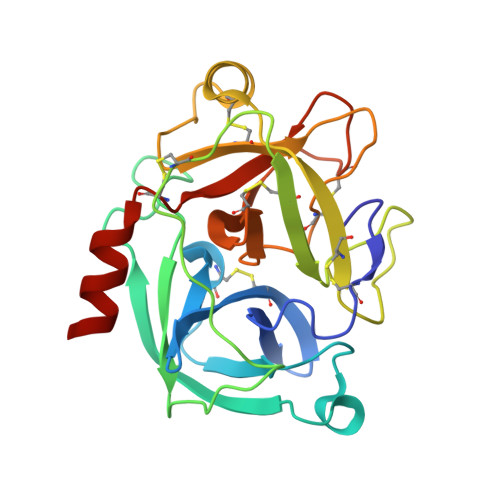
Crystal structure of the inhibitor-free form of the serine protease kallikrein-4.
Riley, B.T., Hoke, D.E., McGowan, S., Buckle, A.M.(2019) Acta Crystallogr F Struct Biol Commun 75: 543-546
- PubMed: 31397325
- DOI: https://doi.org/10.1107/S2053230X19009610
- Primary Citation of Related Structures:
6NVB - PubMed Abstract:
Kallikrein 4 (KLK4) is a serine protease that is predominantly expressed in the prostate and is overexpressed in prostate cancer. As such, it has gained attention as an attractive target for prostate cancer therapeutics. Currently, only liganded structures of KLK4 exist in the Protein Data Bank. Until now, inferences about the subtle structural changes in KLK4 upon ligand binding have been made by comparison to other liganded forms, rather than to an apo form. In this study, an inhibitor-free form of KLK4 was crystallized. The crystals obtained belonged to space group P1, contained four molecules in the asymmetric unit and diffracted to 1.64 Å resolution. Interestingly, a nonstandard rotamer of the specificity-determining residue Asp189 was observed in all chains. This model will provide a useful unliganded structure for the future structure-guided design of KLK4 inhibitors.
Organizational Affiliation:
Biomedicine Discovery Institute and Department of Biochemistry and Molecular Biology, Monash University, 23 Innovation Walk, Clayton, VIC 3800, Australia.