Cryo-EM structures of a human-cockroach hybrid Nav channel in the presence and absence of the alpha-scorpion toxin AaH2.
Clairfeuille, T., Rohou, A., Payandeh, J.(2019) Science
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
(2019) Science
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Sodium channel protein type 9 subunit alpha, Sodium channel protein PaFPC1, chimeric construct | 1,559 | Periplaneta americana, Homo sapiens | Mutation(s): 0 Membrane Entity: Yes | 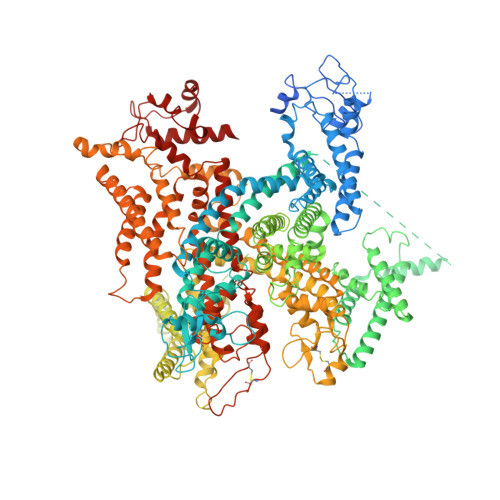 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q15858 (Homo sapiens) Explore Q15858 Go to UniProtKB: Q15858 | |||||
PHAROS: Q15858 GTEx: ENSG00000169432 | |||||
Find proteins for D0E0C2 (Periplaneta americana) Explore D0E0C2 Go to UniProtKB: D0E0C2 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Groups | D0E0C2Q15858 | ||||
Glycosylation | |||||
| Glycosylation Sites: 6 | Go to GlyGen: Q15858-1 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | 2D Diagram | Glycosylation | 3D Interactions |
| beta-D-mannopyranose-(1-3)-[beta-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-2-acetamido-2-deoxy-beta-D-glucopyranose | B | 5 |  | N-Glycosylation | |
Glycosylation Resources | |||||
GlyTouCan: G61217QH GlyCosmos: G61217QH GlyGen: G61217QH | |||||
| Ligands 5 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| AJP Query on AJP | O [auth A] | Digitonin C56 H92 O29 UVYVLBIGDKGWPX-KUAJCENISA-N |  | ||
| 76F Query on 76F | J [auth A], K [auth A], L [auth A], M [auth A] | (7E,21R,24S)-27-amino-24-hydroxy-18,24-dioxo-19,23,25-trioxa-24lambda~5~-phosphaheptacos-7-en-21-yl (9Z,12E)-octadeca-9,12-dienoate C41 H76 N O8 P SUGLKZVXTGKYMP-QNGDKYGQSA-N |  | ||
| LHG Query on LHG | N [auth A] | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE C38 H75 O10 P BIABMEZBCHDPBV-MPQUPPDSSA-N |  | ||
| Y01 Query on Y01 | I [auth A] | CHOLESTEROL HEMISUCCINATE C31 H50 O4 WLNARFZDISHUGS-MIXBDBMTSA-N |  | ||
| NAG Query on NAG | E [auth A], F [auth A], G [auth A], H [auth A] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| Task | Software Package | Version |
|---|---|---|
| RECONSTRUCTION | cisTEM |