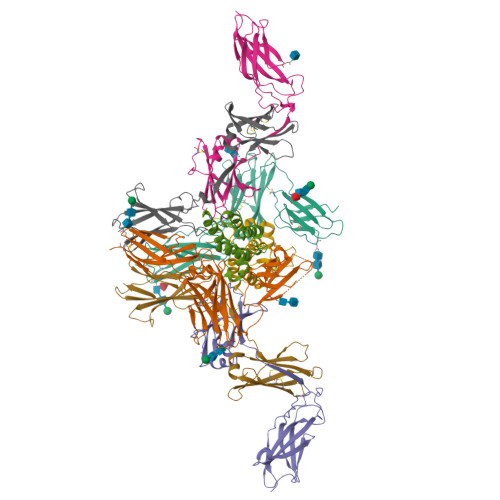
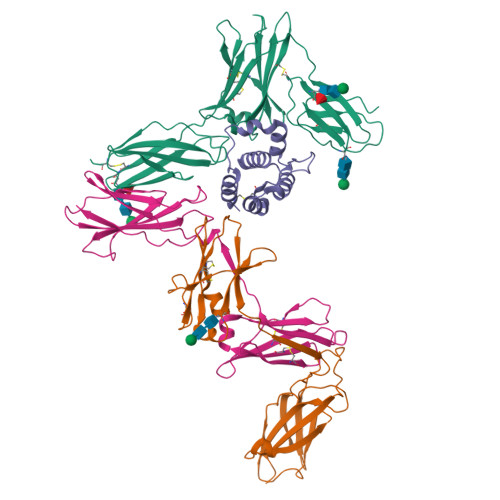
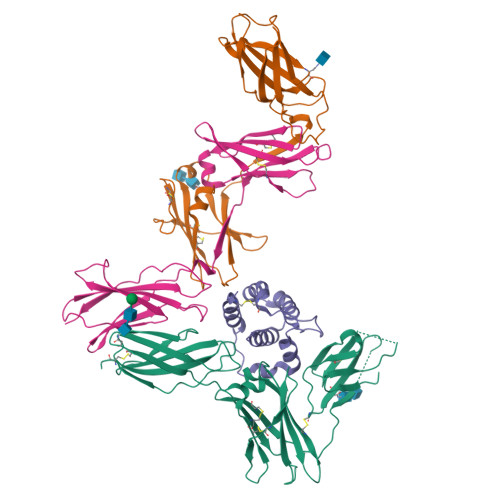
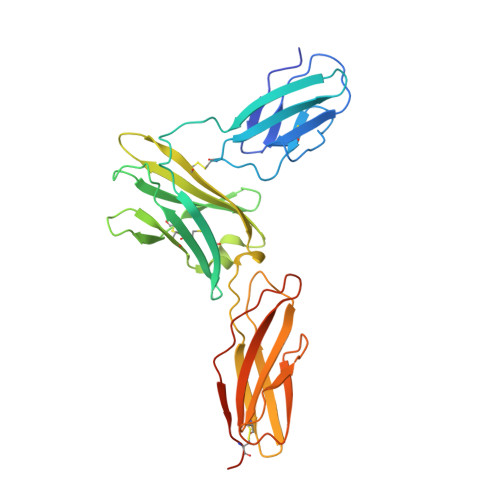
Distinct Assemblies of Heterodimeric Cytokine Receptors Govern Stemness Programs in Leukemia.
Kan, W.L., Dhagat, U., Kaufmann, K.B., Hercus, T.R., Nero, T.L., Zeng, A.G.X., Toubia, J., Barry, E.F., Broughton, S.E., Gomez, G.A., Benard, B.A., Dottore, M., Cheung Tung Shing, K.S., Boutzen, H., Samaraweera, S.E., Simpson, K.J., Jin, L., Goodall, G.J., Begley, C.G., Thomas, D., Ekert, P.G., Tvorogov, D., D'Andrea, R.J., Dick, J.E., Parker, M.W., Lopez, A.F.(2023) Cancer Discov 13: 1922-1947
- PubMed: 37191437
- DOI: https://doi.org/10.1158/2159-8290.CD-22-1396
- Primary Citation of Related Structures:
6NMY - PubMed Abstract:
Leukemia stem cells (LSC) possess distinct self-renewal and arrested differentiation properties that are responsible for disease emergence, therapy failure, and recurrence in acute myeloid leukemia (AML). Despite AML displaying extensive biological and clinical heterogeneity, LSC with high interleukin-3 receptor (IL3R) levels are a constant yet puzzling feature, as this receptor lacks tyrosine kinase activity. Here, we show that the heterodimeric IL3Rα/βc receptor assembles into hexamers and dodecamers through a unique interface in the 3D structure, where high IL3Rα/βc ratios bias hexamer formation. Importantly, receptor stoichiometry is clinically relevant as it varies across the individual cells in the AML hierarchy, in which high IL3Rα/βc ratios in LSCs drive hexamer-mediated stemness programs and poor patient survival, while low ratios mediate differentiation. Our study establishes a new paradigm in which alternative cytokine receptor stoichiometries differentially regulate cell fate, a signaling mechanism that may be generalizable to other transformed cellular hierarchies and of potential therapeutic significance. Stemness is a hallmark of many cancers and is largely responsible for disease emergence, progression, and relapse. Our finding that clinically significant stemness programs in AML are directly regulated by different stoichiometries of cytokine receptors represents a hitherto unexplained mechanism underlying cell-fate decisions in cancer stem cell hierarchies. This article is highlighted in the In This Issue feature, p. 1749.
Organizational Affiliation:
Cytokine Receptor Laboratory, Centre for Cancer Biology, SA Pathology and the University of South Australia, Adelaide, South Australia, Australia.