Fragment-Based Discovery of an Apolipoprotein E4 (apoE4) Stabilizer.
Petros, A.M., Korepanova, A., Jakob, C.G., Qiu, W., Panchal, S.C., Wang, J., Dietrich, J.D., Brewer, J.T., Pohlki, F., Kling, A., Wilcox, K., Lakics, V., Bahnassawy, L., Reinhardt, P., Partha, S.K., Bodelle, P.M., Lake, M., Charych, E.I., Stoll, V.S., Sun, C., Mohler, E.G.(2019) J Med Chem 62: 4120-4130
- PubMed: 30933499
- DOI: https://doi.org/10.1021/acs.jmedchem.9b00178
- Primary Citation of Related Structures:
6NCN, 6NCO - PubMed Abstract:
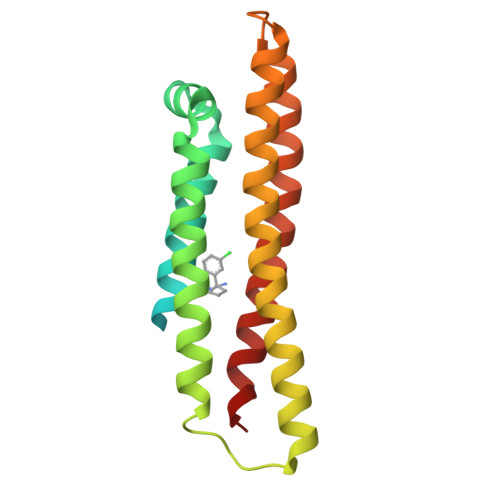
Apolipoprotein E is a 299-residue lipid carrier protein produced in both the liver and the brain. The protein has three major isoforms denoted apoE2, apoE3, and apoE4 which differ at positions 112 and 158 and which occur at different frequencies in the human population. Genome-wide association studies indicate that the possession of two apoE4 alleles is a strong genetic risk factor for late-onset Alzheimer's disease (LOAD). In an attempt to identify a small molecule stabilizer of apoE4 function that may have utility as a therapy for Alzheimer's disease, we carried out an NMR-based fragment screen on the N-terminal domain of apoE4 and identified a benzyl amidine based fragment binder. In addition to NMR, binding was characterized using various other biophysical techniques, and a crystal structure of the bound core was obtained. Core elaboration ultimately yielded a compound that showed activity in an IL-6 and IL-8 cytokine release assay.
Organizational Affiliation:
Research & Development , AbbVie , 1 North Waukegan Road , North Chicago , Illinois 60064 , United States.