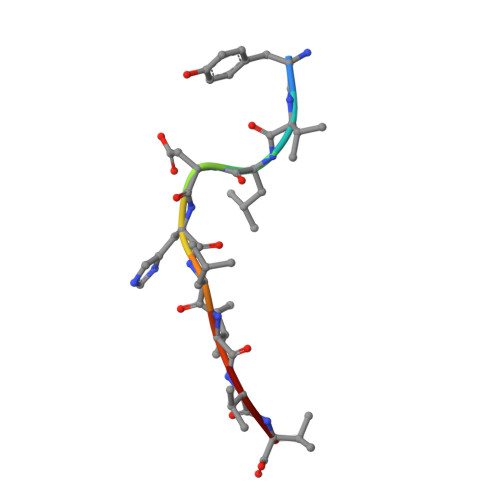
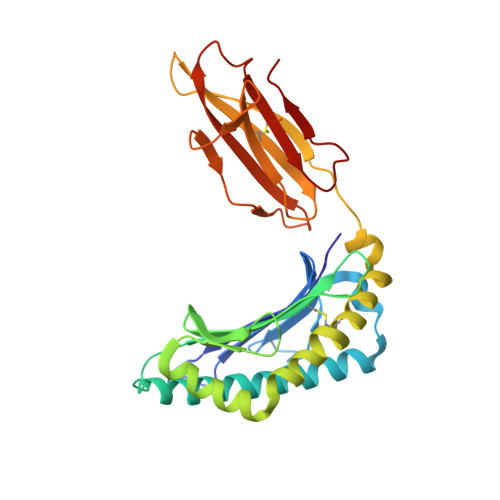
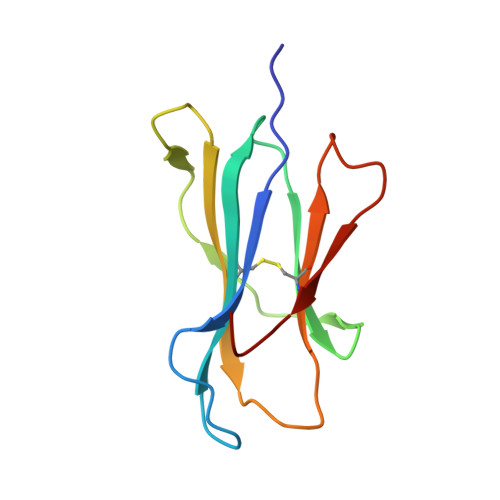
CDR3 alpha drives selection of the immunodominant Epstein Barr virus (EBV) BRLF1-specific CD8 T cell receptor repertoire in primary infection.
Kamga, L., Gil, A., Song, I., Brody, R., Ghersi, D., Aslan, N., Stern, L.J., Selin, L.K., Luzuriaga, K.(2019) PLoS Pathog 15: e1008122-e1008122
- PubMed: 31765434
- DOI: https://doi.org/10.1371/journal.ppat.1008122
- Primary Citation of Related Structures:
6NCA - PubMed Abstract:
The T cell receptor (TCR) repertoire is an essential component of the CD8 T-cell immune response. Here, we seek to investigate factors that drive selection of TCR repertoires specific to the HLA-A2-restricted immunodominant epitope BRLF1109-117 (YVLDHLIVV) over the course of primary Epstein Barr virus (EBV) infection. Using single-cell paired TCRαβ sequencing of tetramer sorted CD8 T cells ex vivo, we show at the clonal level that recognition of the HLA-A2-restricted BRLF1 (YVL-BR, BRLF-1109) epitope is mainly driven by the TCRα chain. For the first time, we identify a CDR3α (complementarity determining region 3 α) motif, KDTDKL, resulting from an obligate AV8.1-AJ34 pairing that was shared by all four individuals studied. This observation coupled with the fact that this public AV8.1-KDTDKL-AJ34 TCR pairs with multiple different TCRβ chains within the same donor (median 4; range: 1-9), suggests that there are some unique structural features of the interaction between the YVL-BR/MHC and the AV8.1-KDTDKL-AJ34 TCR that leads to this high level of selection. Newly developed TCR motif algorithms identified a lysine at position 1 of the CDR3α motif that is highly conserved and likely important for antigen recognition. Crystal structure analysis of the YVL-BR/HLA-A2 complex revealed that the MHC-bound peptide bulges at position 4, exposing a negatively charged aspartic acid that may interact with the positively charged lysine of CDR3α. TCR cloning and site-directed mutagenesis of the CDR3α lysine ablated YVL-BR-tetramer staining and substantially reduced CD69 upregulation on TCR mutant-transduced cells following antigen-specific stimulation. Reduced activation of T cells expressing this CDR3 motif was also observed following exposure to mutated (D4A) peptide. In summary, we show that a highly public TCR repertoire to an immunodominant epitope of a common human virus is almost completely selected on the basis of CDR3α and provide a likely structural basis for the selection. These studies emphasize the importance of examining TCRα, as well as TCRβ, in understanding the CD8 T cell receptor repertoire.
Organizational Affiliation:
Program in Molecular Medicine, University of Massachusetts Medical School, Worcester, Massachusetts, United States of America.